Abstract
The Atlantic–Mediterranean area has recently been proposed as a new phylogeographical area on the basis of concordance of genetic differentiation patterns observed in several marine species. However, additional taxa need to be studied to establish the phylogeographical relationship between the Atlantic and Mediterranean. Eleven samples of the cuttlefish Sepia officinalis around the Iberian Peninsula, one from the Canary Islands, and another from Fiumicino (Italy) were screened for 33 allozyme loci. Genetic variability was low in all samples (He between 0.022 and 0.076). Intersample genetic differentiation was high (FST=0.220), mainly because of genetic variation in the non-Iberian samples. One locus (PEPD*), diagnostic between the Italian sample and all others, suggests the possible existence of hitherto unrecognized species or subspecies of Sepia in the Mediterranean Sea. The 11 Iberian samples exhibited moderate genetic differentiation (FST=0.100), which could be explained on the basis of genetic differentiation between Atlantic and Mediterranean samples. Significant clines in allele frequencies were observed for five out of six polymorphic loci. These results support a model of secondary intergradation (i.e. secondary contact of populations that were previously differentiated in isolation) similar to that previously proposed for other marine species from the Atlantic–Mediterranean area.
Similar content being viewed by others
Introduction
Studies on several different marine fishes and molluscs (see Sanjuan et al., 1996a, 1997 and references therein; Roldán et al., 1998) have revealed geographically concordant intersample genetic differentiation between Atlantic and Mediterranean samples. A hypothesis has been proposed to explain these patterns, wherein Mediterranean populations were isolated from Atlantic populations during ice-ages (Pliocene and Quaternary), with subsequent genetic divergence, post ice-age secondary contact, and present-day gene flow between populations restricted by the Straits of Gibraltar or the Almería-Oran oceanographic front. Nevertheless, additional studies using taxa with different biological characteristics are needed to clarify and confirm the importance of this region as a new phylogeographical area.
Sepia officinalis (Linnaeus 1758; Cephalopoda: Sepiidae) is an important fishery resource for European and North African countries (annual captures are around 70 000 metric tonnes; FAO, 1994). It is distributed along the NE Atlantic continental margin, from the Baltic Sea to Senegal, and throughout the Mediterranean Sea (Guerra, 1992). This cephalopod fixes its eggs to the sea floor, a larval phase is lacking, and the adults have limited migratory capacity, so its dispersive ability is presumed to be restricted (Guerra, 1992).
Allozyme polymorphisms have proved to be effective for estimating population divergence and identifying discrete fish and cephalopod stocks (Carvalho & Hauser, 1994 and references therein). The one previous allozyme-based study on Sepia officinalis (Sanjuan et al., 1996b) found no significant genetic differences between samples on either side of a subsurface oceanographic front off the NW Iberian Peninsula. The aim of the present study was to analyse the genetic structure, using allozyme markers, of S. officinalis populations from the Atlantic and Mediterranean Sea for geographical patterns of genetic variation.
Materials and methods
Sampling
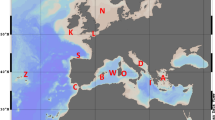
Thirteen samples of Sepia officinalis were collected from commercial catches in NE Atlantic and Mediterranean fishing ports between March 1993 and November 1997 (Table 1 and Fig. 1). Eleven samples were caught from waters around the Iberian Peninsula, six from Atlantic waters (RoBo, Dono, GalN, GalW, FFoz and SBar), and five from the Mediterranean Sea (CVel, RMar, CPla, VGel and Rosa); one sample was taken off the Canary Islands (ICan); and another off Fiumicino (Fium), Italy. The specimens were obtained on the date of capture and were immediately frozen in dry ice and stored at −72°C until required.
Samples of Sepia officinalis collected in fishing ports along the Atlantic and Mediterranean coasts. Sample codes are indicated in Table 1.
Electrophoresis
Samples of mantle muscle were prepared for electrophoresis using methods previously described for Sepia species (Pérez-Losada et al., 1996). Standard horizontal starch gel electrophoresis was carried out (Murphy et al., 1996). Twenty-eight enzymes, yielding 33 putative enzyme coding loci, displayed adequate activity and resolution for consistent interpretation and routine examination. Detailed electrophoretic conditions and histochemical staining recipes for most of the enzymes are described in Pérez-Losada et al. (1996). For ALPDH, G6PDH and GAPDH the electrode buffer was Tris-Citrate pH 8.0 (gel buffer dilution 1:9), and histochemical staining recipes were as in Murphy et al. (1996).
Data analysis
Genotype frequencies at polymorphic loci were tested for agreement with Hardy–Weinberg equilibrium expectations by chi-squared tests, and the probability of the null hypothesis was estimated using Monte Carlo simulation. The genetic structure of samples was analysed by means of F-statistics (Nei, 1987). The significance of FST was calculated by a chi-squared test of homogeneity of allele frequencies and the probability of the null hypothesis was estimated using Monte Carlo simulation. Mean expected heterozygosity per locus (unbiased estimate), observed heterozygosity, mean number of alleles and proportion of polymorphic loci (Nei, 1987) were calculated for each sample.
Nei’s (1978) (DN) and Cavalli-Sforza & Edwards’s (1967) arc (Darc) genetic distances among samples were computed. These values were then used to construct UPGMA (unweighted pair-group method using arithmetic averages; Sneath & Sokal, 1973) and neighbour-joining (Saitou & Nei, 1987) trees.
The correlation coefficient between the matrix of Cavalli-Sforza & Edwards’s (1967) arc genetic distances and geographical distances was calculated and its probability was estimated by means of a Mantel test based on 1000 permutations. Allele frequencies of the most polymorphic loci were plotted against geographical distances among samples, and the significance of allozyme variation patterns was tested by correlation coefficients between the arcsine-squared-root-transformed allele frequencies and geographical distances. The sequential Bonferroni technique (Rice, 1989) was used to adjust significance levels for multiple simultaneous comparisons.
BIOSYS-1 (Swofford & Selander, 1981) and Zaykin & Pudovkin (1993) computer programs were used to perform most of the genetic analyses. The Mantel test was carried out using the NTSYS-PC computer program (Rohlf, 1994).
Results
Genetic variability
Seventeen enzyme loci were monomorphic in all samples; allele frequencies at the 16 polymorphic loci are shown in Table 2Table 2. Eight loci showed one or two rare alleles (allele frequency <0.05), and seven loci exhibited a moderate variability with two or three alleles per locus (AAT-1*, ESTD*, IDDH*, IDHP*, MEP*, OPDH-1* and PGDH*).
The PEPD* locus was diagnostic between the Fium sample (N=24) and all 12 other Sepia samples (N=328). In addition, at locus AAT-1* the Italian sample had an AAT-1*100 allele frequency of only 0.396, whereas in the other 12 samples this allele was nearly fixed. The IDDH* locus showed a fixed allele (IDDH*85) in the ICan sample, whereas this allele varied from 0.100 (VGel and Rosa) to 0.667 (RoBo) in the other 12 samples.
The F-values between loci and samples showed great variability, although no significant deviations from Hardy–Weinberg expectations were found. The estimates of genetic variability for all 13 S. officinalis samples (Table 2) were low for all indices (He ranged between 0.022 and 0.076) and the He over all samples was 0.057 ± 0.022. Mediterranean samples exhibited the highest values for the variability indices, although He was not significantly different between Mediterranean and Atlantic samples [Student’s t11=0.710, 0.40 < P < 0.50; Nei (1987)].
Population structure
The mean FST value over all 13 samples was 0.220. This high value of genetic differentiation was mainly caused by allele frequencies in Fium for PEPD* and AAT-1*, and ICan for IDDH* (see Table 2). When both samples were excluded from the analysis, the mean FST over all 11 Iberian samples was halved (FST=0.100, Table 3). All the most polymorphic loci (P95) except AAT-1*, plus ALPDH* and OPDH-2*, exhibited significant heterogeneity in allele frequencies among Iberian samples (Table 3).
Genetic distances of Nei (1978) (DN) and Cavalli-Sforza & Edwards (1967) (Darc) are presented in Table 4. Both distances showed relatively low values for all pairwise comparisons; DN ranges between 0.000 and 0.056, and Darc between 0.024 and 0.238. Maximum distances occur between the geographically most distant samples (Fium vs. RoBo or Dono). The UPGMA trees (Fig. 2) showed that the non-Iberian samples (Fium and ICan) were clearly separated from the Iberian samples, which fell into three groups. The most northern Iberian samples (RoBo and Dono) were clustered and separated from the east Iberian samples (CPla, VGel and Rosa), which constituted another group. In the UPGMA based on DN(Fig. 2a) the six geographically intermediate Atlantic and Mediterranean samples were clustered together. However, in the UPGMA based on Darc (Fig. 2b), these intermediate samples were split into western and southern clusters, except RMar, which was joined to the western group. When the neighbour-joining method was applied to both distance matrices, trees with the same topology as those presented in Fig. 2 were obtained. Genetic distances were significantly correlated with geographical distances between samples (r=0.721, P < 0.001; Fig. 3), but genetic distances were greater than expected for the 12 Fium comparisons.
UPGMA dendrograms showing relationships among 13 samples of Sepia officinalis based on Nei’s (1978) and Cavalli-Sforza & Edwards’s (1967) arc genetic distances. The cophenetic correlations were ra=0.982 and rb=0.970. Sample codes are indicated in Table 1. N, the most northern Iberian samples; W, north-western and western Iberian samples; S and E, southern and eastern Iberian samples, respectively.
Correlation between Cavalli-Sforza & Edwards’s (1967) arc genetic distance and geographical distance (km) for 13 samples of Sepia officinalis. o: Fium with every one of the other 12 samples. r=0.721, P < 0.001 (Mantel test).
Separation of the 11 Iberian samples into Atlantic and Mediterranean regional groups is further supported by consideration of variation over all loci. In the Atlantic Region (AR) were grouped RoBo, Dono, GalN, GalW and FFoz; and in the Mediterranean Region (MR) were included SBar, CVel, RMar, CPla, VGel and Rosa (Table 3). SBar was considered as a Mediterranean sample on the basis of its geographical proximity to the Mediterranean Sea and because it is genetically more similar to Mediterranean samples (Fig. 2b). The mean genetic differentiation (FST, Table 3) decreased from 0.100 over all Iberian collections to 0.063 for AR, and to 0.061 for MR, the decrease being specially important for some of the polymorphic loci such as ESTD* and MEP*.
Clearly significant geographical allozyme variation appeared for three loci (after the application of the Bonferroni test) with a moderate genetic variability: IDDH*, MEP* and OPDH-1* (Fig. 4). In the case of MEP*, RMar behaved as an outlier for MEP*75, showing an allele frequency of 0.094. Slight but significant allozyme variation was also observed at two loci with low levels of genetic variability: ESTD* and OPDH-2*. A V-shaped pattern of variation was found at the IDHP* locus which showed significant clines on both sides of the Straits of Gibraltar, but not around the Iberian Peninsula as a whole.
Allele frequency variation at polymorphic loci ESTD*, IDDH*, IDHP*, MEP*, OPDH-1* and OPDH-2* for 11 Iberian samples of Sepia officinalis against geographical distance. RoBo is at 0 km and Rosa is at 31 km (× 100). All loci, except IDHP*, displayed significant geographical allozyme variation around the Iberian Peninsula.
Discussion
The study of 33 enzyme loci in 13 samples of S. officinalis showed that this species has low levels of allozyme variability; the mean He (0.057 ± 0.022) falls below the average for invertebrates and molluscs (0.122 and 0.145, respectively; Ward et al., 1992), and also for marine molluscs (0.147; Fujio et al., 1983), but within the range for cephalopods (see Brierley et al., 1996; Sanjuan et al., 1996b; and references therein).
Despite the observed low genetic variability, extensive intersample variation (FST=0.220) was observed over all the samples studied. This genetic differentiation could result from the allozyme differences between the non-Iberian samples (Fium and ICan) and the Iberian samples, and the genetic differences between the samples considered as Atlantic (AR) and Mediterranean (MR). The Tyrrhenian sample (Fium) was clearly separated from the other samples in the UPGMA trees (Fig. 2). The genetic differentiation of this sample can be attributed mainly to the diagnostic locus PEPD* (FST=1.000) and, to a lesser degree, to the locus AAT-1* (FST=0.565). These results suggest that the individuals from Fium are genetically distinct (no gene flow between these and the other samples), and may even comprise a separate and hitherto unrecognized subspecies or species of Sepia. Both Nesis (1987) and Khromov (pers. comm.) have suggested that, based on morphological data, the notion of a subspecies of S. officinalis in the Mediterranean Sea (S. officinalis mediterranea Nini 1884) is valid. Nevertheless, the unbiased genetic identity (I; Nei, 1978) between Fium and the other 12 samples (0.954–0.946) is of the order generally considered to be indicative of conspecific populations (Thorpe, 1983). A more extensive study combining molecular (allozyme and DNA) markers with morphological characters, and more samples between Rosa and Fium will be needed to clarify this hypothesis.
The Canary Islands sample (ICan) is also distinctly divergent from the other samples in the UPGMA tree (Fig. 2), mainly because of genetic variation at the loci IDDH*, MEP* and OPDH-1*. This sample is most closely related genetically (DN=0.008; Darc=0.106) to the SBar sample, which is also the geographically most proximal (≈ 1000 km). Thus, isolation by distance could be responsible for the genetic divergence of this sample from the Iberian samples.
The Iberian samples showed moderate intersample differences overall (FST=0.100; Table 3), although genetic distances for all pairwise comparisons were low (DN < 0.03; Table 4). Genetic differences are significantly correlated with increasing geographical separation along the coastline between the Atlantic and Mediterranean (Fig. 3). These results are supported by the clustering of samples (Fig. 2), which match rather well their geographical distribution. The UPGMA tree based on Darc (Fig. 2b) suggests a separation of the samples from the Atlantic Region (AR) from those of the Mediterranean Region (MR). In the same way, the genetic differentiation between Atlantic and Mediterranean samples is also evident in the hierarchical analysis of F-statistics (Table 3), which indicates greater genetic heterogeneity between than within regions. Similarly, the estimate of number of effective migrants (Nm, where Nm=[1/FST – 1]/4) over all 11 Iberian samples (Nm=2.3) reflects a moderate amount of gene flow, and was lower than Nm for each region separately (Nm=3.7 for AR; Nm=3.9 for MR). Samples from the MR possessed nine alleles exclusive to this area, whereas only three alleles were exclusive to AR. This difference in number of exclusive alleles may be an indication of a real barrier to gene flow outwards from the Mediterranean Sea, and suggests that alleles originating in Mediterranean populations are prevented from spreading to Atlantic populations, whereas those arising in Atlantic populations are spreading into the Mediterranean Sea. This asymmetrical gene flow could be maintained by the unidirectional marine surface circulation southwards along the coast of the Iberian Peninsula and into the Mediterranean Sea across the Straits of Gibraltar (Rodríguez, 1982).
This study also showed that geographical allozyme variation around the Iberian Peninsula is a major feature of allozyme intersample variability in S. officinalis (Fig. 4). Such variation in allele frequencies may be the result of secondary contact of cuttlefish populations that were isolated in the past and became differentiated in allopatry (i.e. secondary intergradation; Endler, 1977; Avise, 1994). In this sense, the main change for allele frequencies roughly coincides with the area of transition from the Atlantic to the Mediterranean, the Straits of Gibraltar and the Alborán Sea, suggesting the position of the barrier that produced such isolation in the past. This transition area is also suggested by the different clustering of the three southern Iberian samples (SBar, CVel and RMar) in the UPGMA trees (Fig. 2), indicating a genetic character intermediate between the geographically distant Atlantic (RoBo–FFoz) and Mediterranean (Cpla–Rosa) samples. This is compatible with what is known of the historical hydrography of this area: intermittent physical barriers, precluding colonization and migration, existed as recently as one million years ago (Maldonado, 1985 and references therein); recent lowering of sea level by 100–200 m during Quaternary glaciations resulted in the partial or total closure of the Straits (Bianco, 1990). One polymorphic locus (IDHP*), however, displayed a V-shaped pattern over the Iberian Peninsula (Fig. 4). The coincidence of the V midpoint at this locus with the transition area described earlier, gives further support to the same geographical separation hypothesis. However, V-shaped patterns cannot arise from the contact of two genetically differentiated populations, and clines implied in these patterns must have originated independently in each sea basin.
The results presented for S. officinalis in this paper support the importance of the Atlantic–Mediterranean area in phylogeographical structuring within marine species (Sanjuan et al., 1996a), as has been similarly suggested for the Gulf of Mexico/Atlantic coasts of North America and the Indian Ocean/Malayan Provinces (Avise, 1994; Palumbi, 1994).
References
Avise, J. C. (1994) Molecular Markers, Natural History and Evolution. Chapman & Hall, New York.
Bianco, P. G. (1990). Potential role of the palaeohistory of the Mediterranean and Paratethys basins on the early dispersal of Euro-Mediterranean freshwater fishes. Ichthyol Expl Freshw, 1: 167–184.
Brierley, A. S., Allcock, A. L., Thorpe, J. P. and Clarke, M. R. (1996). Biochemical genetic evidence supporting the taxonomic separation of Loligo edulis and Loligo chinensis (Cephalopoda: Teuthoidea) from the genus Loligo. Mar Biol, 127: 97–104.
Carvalho, G. R. and Hauser, L. (1994). Molecular genetics and the stock concept in fisheries. Rev Fish Biol Fish, 4: 300–326.
Cavalli-Sforza, L. L. and Edwards, A. W. F. (1967). Phylogenetic analysis: models and estimation procedures. Evolution, 32: 550–570.
Endler, J. A. (1977) Geographic Variation, Speciation, and Clines. Princeton University Press, Princeton, NJ.
FAO (FOOD AND AGRICULTURE ORGANIZATION) (1994) Yearbook Fishery Statistics 1992 no. 74. FAO, Rome.
Fujio, Y., Yamanaka, R. and Smith, P. J. (1983). Genetic variation in marine molluscs. Bull Jap Soc Sci Fish, 49: 1809–1817.
Guerra, A. (1992). Mollusca, Cephalopoda. In: Ramos, M. A. et al(eds) Fauna Ibérica vol. 1. Museo Nacional de Ciencias Naturales, CSIC, Madrid.
Maldonado, A. (1985). Evolution of the Mediterranean basins and a reconstruction of the Cenozoic palaeoceanography. In: Margalef, R. (ed.) Western Mediterranean, pp. 18–61. Pergamon Press, London.
Murphy, R. W., Sites, C. W. Jr, Buth, D. G. and Haufler, C. H. (1996). Proteins: isozyme electrophoresis. In: Hillis, D. M. and Moritz, C. (eds) Molecular Systematics. pp. 51–120. Sinauer Associates, Sunderland, MA.
Nei, M. (1978). Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics, 89: 583–590.
Nei, M. (1987) Molecular Evolutionary Genetics. Columbia University Press, New York.
Nesis, K. N. (1987) Cephalopods of the World. T. H. F. Publications, Neptune City, NJ.
Palumbi, S. R. (1994). Genetic divergence, reproductive isolation, and marine speciation. Ann Rev Ecol Syst, 25: 547–572.
Pérez-Losada, M., Guerra, A. and Sanjuan, A. (1996). Allozyme electrophoretic technique and phylogenetic relationships in three species of Sepia (Cephalopoda: Sepiidae). Comp Biochem Physiol B, 114: 11–18.
Rice, W. R. (1989). Analyzing tables of statistical tests. Evolution, 43: 223–225.
Rodríguez, J. (1982) Oceanografía Del Mar Mediterráneo. Pirámide, Madrid.
Rohlf, F. J. (1994) Ntsys-Pc. Numerical Taxonomy and Multivariate Analysis System Version 1.80. Exeter Software, New York, NY.
Roldán, M. I., García-Marín, J. L., Utter, F. M. and Pla, C. (1998). Population genetic structure of European hake, Merluccius merluccius. Heredity, 81: 327–334.
Saitou, N. and Nei, M. (1987). The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol, 4: 406–425.
Sanjuan, A., Comesaña, S. and De Carlos, A. (1996a). Macrogeographic differentiation by mtDNA restriction site analysis in the SW European Mytilus galloprovincialis Lmk. J Exp Mar Biol Ecol, 198: 89–100.
Sanjuan, A., Pérez-Losada, M. and Guerra, A. (1996b). Genetic differentiation in three Sepia species (Mollusca: Cephalopoda) from Galician waters (Northwest Iberian Peninsula). Mar Biol, 126: 253–259.
Sanjuan, A., Zapata, C. and Álvarez, G. (1997). Genetic differentiation in Mytilus galloprovincialis Lmk. throughout the world. Ophelia, 47: 13–31.
Sneath, P. H. and Sokal, R. R. (1973) Numerical Taxonomy. W. H. Freeman, San Francisco.
Swofford, D. L. and Selander, R. B. (1981). Biosys-1: a Fortran program for the comprehensive analysis of electrophoretic data in population genetics and systematics. J Hered, 72: 281–283.
Thorpe, J. P. (1983). Enzyme variation, genetic distance and evolutionary divergence in relation to levels of taxonomic speciation. In: Oxford, G. S. and Rollinson, D. (eds) Protein Polymorphism. Adaptive and Taxonomic Significance. pp. 131–152. Academic Press, London.
Ward, R. D., Skibinski, D. O. F. and Woodwark, M. (1992). Protein heterozygosity, protein structure and taxonomic differentiation. Evol Biol, 26: 73–159.
Zaykin, D. V. and Pudovkin, A. I. (1993). Two programs to estimate significance of χ2values using pseudo-probability tests. J Hered, 84: 152–152.
Acknowledgements
We are grateful to P. Shaw for his valuable suggestions on a draft of the manuscript and two anonymous referees for their interesting comments. We thank F. Casas, J. M. Olveira, L. Pozzan, J. Ferrer, M. Bernal and A. Rodrigues for assistance in the collection of the samples. Thanks are also due to A. Lavalatina for her MAC care. This research was supported by the AMB94–0371 project (CICYT, Spain) and its complementary one 64102C503 (University of Vigo, Spain) to A.S. This paper fulfils the Ph.D. requirements for M.P.-L., who received a Scholarship from Xunta de Galicia (Spain).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Pérez-Losada, M., Guerra, Á. & Sanjuan, A. Allozyme differentiation in the cuttlefish Sepia officinalis (Mollusca: Cephalopoda) from the NE Atlantic and Mediterranean. Heredity 83, 280–289 (1999). https://doi.org/10.1038/sj.hdy.6885520
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6885520
Keywords
This article is cited by
-
Genetic diversity and population structure of Sepia officinalis from the Tunisian cost revealed by mitochondrial COI sequences
Molecular Biology Reports (2015)
-
Tropical fishes in a temperate sea: evolution of the wrasse Thalassoma pavo and the parrotfish Sparisoma cretense in the Mediterranean and the adjacent Macaronesian and Cape Verde Archipelagos
Marine Biology (2008)
-
Approaches to resolving cephalopod movement and migration patterns
Reviews in Fish Biology and Fisheries (2007)
-
Phylogeography and evolution of the triplefin Tripterygion delaisi (Pisces, Blennioidei)
Marine Biology (2007)
-
Phylogeographic patterns of the mysid Mesopodopsis slabberi (Crustacea, Mysida) in Western Europe: evidence for high molecular diversity and cryptic speciation
Marine Biology (2006)