Abstract
Lolium perenne (4x)/ Festuca pratensis (2x) triploid hybrids (2n=3x=21) were produced and backcrossed to Lolium perenne. The BC1 progeny, which predominantly had 14 chromosomes, were analysed using genomic in situ hybridization (GISH) and genetic markers. GISH revealed that over 74% of the BC1 individuals carried one or more F. pratensis chromosome segments. By comparing the physical size of introgressed F. pratensis chromosome segments with the presence or absence of F. pratensis-specific polymorphisms, it was possible to determine the physical position of genetic markers. The potential of a new type of genetic mapping (‘introgression mapping’) for the alignment of physical and genetic maps, determining the genetic control of agronomically important characters and the production of novel germplasm for the development of new varieties is discussed.
Similar content being viewed by others
Introduction
Genome mapping is a powerful approach to the study of plant biology, facilitating the location of genes responsible for agronomically and scientifically important traits. Phenotype mapping is the primary justification for the development of genetic maps. DNA markers closely linked to a gene responsible for a desirable phenotype are an important tool for indirect selection in breeding programmes and in addition provide a springboard from which genes can be isolated via map-based cloning. To increase the efficiency of mapping, parental genotypes which exhibit high DNA marker allele diversity are chosen. The genetic base of many crop species, however, is narrow, but this problem can be overcome by generating maps from wide crosses.
The forage grass Lolium perenne (Lp) (2n=2x=14) can be readily hybridized with Festuca pratensis (Fp) (2n=2x=14) to form a 14-chromosome hybrid which exhibits full pairing at metaphase I of meiosis (Lewis, 1966; Jauhar, 1975). These diploid hybrids show nearly complete sterility (Lewis, 1966; Jauhar, 1975). However, LpLpFp triploids, derived by hybridizing synthetic tetraploid L. perenne with diploid F. pratensis, show both male and female fertility (Jauhar, 1975). The ease of hybridization, the chromosome pairing observed at meiosis and a preliminary molecular genetic study on the phylogeny of these species (Stammers et al., 1995) indicate that they are closely related. In contrast, the sterility of the diploid hybrid, the ease with which Lp and Fp chromosomes can be distinguished using genomic in situ hybridization (GISH) (King et al., 1997), and the position and number of rDNA sites in the two species (Thomas et al., 1996; 1997) indicates that they are distantly related. Thus the phylogenetic relationship between Lp and Fp at present remains a paradox.
The work presented in this paper describes a new kind of genetic mapping in the grasses. ‘Introgression mapping’ is based on the ability to distinguish Lp and Fp chromosomes and chromosome segments visually using GISH. The potential is discussed of introgression mapping for the development and alignment of physical and genetic maps, determining the genetic control of agronomically important characters and the production of novel germplasm for the development of new varieties.
Materials and methods
Plant material
Triploid LpLpFp hybrids were generated by intercrossing the synthetic autotetraploid Lp (variety Meltra, 2n=4x=28), as the female parent, with diploid Fp (Bf 1183, 2n=2x=14) as the pollen parent. A single LpLpFp triploid hybrid (2n=3x=21) resulting from this cross was then used as the pollen parent in crosses to a single diploid Lp genotype (variety Liprio). From a BC1 population of 161 plants an initial sample of 78 were selected and screened using GISH. These 78 plants were also characterized with genetic markers, i.e. AFLPs, RFLPs and RAPDs.
Plants identified as carrying a single Fp chromosome using GISH were backcrossed again to Lp (variety Liprio) to produce a BC2 population.
In situ hybridization
Total genomic DNA was extracted from young actively growing leaves of Fp and Lp as described by Dellaporta et al. (1983). Fp DNA which was used exclusively as the probe was sheared to ≈5 kb using a sonicating water bath and then labelled using either digoxigenin-11-dUTP (Boehringer-Mannheim), rhodamine-4-dUTP (Amersham) or Texas Red-5-dCTP (Dupont). Blocking DNA was prepared by autoclaving the DNA of Lp for 2 min, which produced fragments of ≈200 bp in length. The in situ hybridization protocol used followed that described by Thomas et al. (1994).
Slides were analysed using a Leica DM/RB epifluorescence microscope with filter blocks for DAPI, fluorescein and rhodamine. Images were captured with a monochrome CCD camera as described by Thomas et al. (1996).
AFLP analysis
AFLP reactions were carried out as described by Vos et al. (1995) using adapters, nonselective and selective primers obtained from Keygene. Briefly, 2.5 μg of genomic DNA was digested with 5 U/μg of EcoRI and MseI and ligated with EcoRI and MseI adapters. The biotin subtraction step was omitted and the ligated templates were preamplified with E01 and M01 primers. Parents and BC1 genotypes were prescreened with combinations of selective primers, and selective amplifications were carried out on the full family with 10 different primer combinations (E35 with M32, M35, M36, M37, M38, M39, M41, M42, M44, and E36 with M36). AFLP bands were visualized either by including 1% digoxigenin-11-dUTP in the selective amplification reactions, transferring to Hybond N (Amersham) hybridization membrane using a Hoefer GeneSweep TE90 electroblotter and detecting with antidig antibodies conjugated with alkaline phosphatase and the chemiluminescent substrate CSPD according to manufacturers' instructions (Boehringer-Mannheim), or by silver staining with the Silver Sequence DNA Staining Reagents (Promega).
RFLP and RAPD analysis
Southern hybridizations, RFLP and RAPD analyses were carried out as in Pessino et al. (1997), except that 1 U of ‘Red Hot’ DNA Polymerase (Advanced Biotechnologies) was used for RAPD amplifications in place of Taq DNA Polymerase (BRL).
Results
Of the 78 BC1 individuals analysed by GISH, 75 had 14 chromosomes and three had 15. Fifty-eight plants, including the three with 15 chromosomes, carried one or more Fp chromosomes or chromosome segments. Three lines carried Lp/Fp chromosome substitutions and these were fully fertile near-normal plants, suggesting the substitution of a homoeologous chromosome. Fourteen plants carried a single Fp chromosome segment which ranged in size from a small segment to more than a whole chromosome arm. Thirty-one plants had two Fp chromosome segments, nine had three and one had six. The remaining 20 plants analysed had no Fp chromosome segment detectable by GISH.
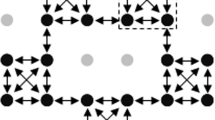
Analysis of the Lp/Fp recombinant chromosomes revealed that homoeologous recombination in the parental LpLpFp triploid plant appeared to occur along the entire length of the Lp and Fp chromosomes (Fig. 1). Some Lp/Fp recombinant chromosomes demonstrated that double-recombinant events occurred between Lp and Fp homoeologous arms (Fig. 1).
Mitoses of Lolium perenne/Festuca pratensis (Lp/Fp) introgression lines with sites of Lp/Fp recombination indicated by arrows: (a) two distal recombination points; (b) an introgression in two different chromosomes, one of which is the result of a double recombinant event; (c) one introgression including the Fp centromere with recombination in both chromosome arms; (d) introgressions into three chromosomes; (e) in addition to the introgressed Fp segment, arrowheads indicate regions of Lp chromosomes which routinely show cross-hybridization with Fp probe (nucleolar organizer regions) (bar=10 μm); (f) shows a series of different lines either carrying a complete Fp chromosome or a single Fp chromosome segment derived from the same chromosome. The physical position of the genetic markers can be determined by the presence or absence of Fp-specific polymorphisms. Determination of the exact order of markers 3538.11 and 3542.17; OPAGO26 and 3536.15; and 3544.2, 3537.16 and 3535A would require analysis of further recombinants or the production of a Lp/Fp genetic map (see Discussion).
AFLPs, RFLPs and RAPDs were used to determine which linkage group each BC1 individual represented. From these data three linkage groups were identified as having whole Fp chromosome substitutions, and two linkage groups were identified as including individuals with single chromosome-arm substitutions. The size of the Fp chromosome segments, determined by GISH, was related to the presence or absence of Fp- specific polymorphisms (Fig. 1f). This determined the approximate physical position of marker loci.
Discussion
The work demonstrates that Lp and Fp chromosomes recombine at high frequency in the LpLpFp triploid hybrids, and that it is possible to distinguish the products of recombination visually by using GISH, as well as by genetic markers. Recombination appears to occur freely along the whole length of the chromosomes. Furthermore, an initial analysis of metaphase I of meiosis in the LpLpFp triploid indicates that the seven Lp chromosomes pair with their Fp homoeologues at equal frequencies (I. P. King submitted).
The ease with which Lp and Fp can be hybridized, the high frequency of homoeologous recombination and the ability to distinguish the products of meiosis visually using GISH makes these species unique among crop plants. The introgression of agronomically important genes into cereals from their distant relatives has been a major goal for geneticists because it enlarges the gene pool available for selection by breeders. Taking wheat (Triticum aestivum) as an example, it is possible to make hybrids with closely related species that carry a common genome (T. urartu A-genome and Aegilops squarrosa D-genome; for review see Gale & Miller, 1987). The chromosomes from the common genomes recombine freely, but it is not possible to distinguish them using GISH.
In more distantly related species whose chromosomes can be distinguished from those of wheat by GISH, wheat/alien recombination does not normally occur because of the action of the pairing control gene Ph1 located on the long arm of chromosome 5B (for review see Gale & Miller, 1987). In the absence of Ph1, wheat/alien recombination occurs but it is at a very much lower frequency than that observed in the LpLpFp hybrids. An example is the wheat/rye hybrids studied by Miller et al. (1994).
Introgression mapping
The combination of promiscuous homoeologous recombination in LpLpFp hybrids and their derivatives, and the ability to distinguish the Lp and Fp genomes visually, provides us with a novel opportunity to develop ‘introgression maps’. The first stage is the isolation of 14 Lp/Fp chromosome-arm substitution lines. In each, a single pair of arms is replaced by the homoeologous Fp arms.
Each line is then crossed to Lp and the F1 backcrossed to generate a series of plants which contain a spectrum of Fp segment sizes in their complementary Lp backgrounds (Fig. 2).
In our experiment where only 78 individuals were analysed, we have established two chromosome-arm substitutions and three whole-chromosome substitutions. If the Lp chromosomes and chromosome arms had been substituted with nonhomoeologous Fp chromosome arms it would be expected that the plants would be inviable or alternatively show retarded growth and low fertility, because Lp is a diploid and not buffered by polyploidy. However, the plants were fully fertile and vigorous. From pairing data there is no evidence for translocation differences between Lp and Fp (unpublished data). Thus it should be possible to isolate the remaining chromosome-arm substitutions from additional BC1 progeny.
The development of the chromosome-arm substitution lines will provide invaluable tools for determining the genetic control of agronomically important characters. They will enable screening to determine which chromosome arms carry genes affecting particular characters. The genes can then be genetically and physically mapped by analysing a recombinant chromosome series (Fig. 2). This approach has similarities to the intervarietal substitution lines developed in wheat (for review see Law et al., 1987), but the Lp/Fp system offers much higher resolution as it is possible to assign genes to a specific location on a chromosome arm.
Although chiasmata may occur anywhere along a chromosome, they are not evenly distributed. In wheat, for example, chiasmata tend to form towards the ends of the chromosomes. Thus it is clear that the genetic distance in cM does not directly equate to the physical distance in base-pairs. The relationship between genetic distance and physical distance is of great importance for map-based cloning strategies, because the effort involved in techniques such as chromosome walking depends on the length of the DNA in question.
There are a number of ways in which the physical distance between genetic markers can be determined in large-genome cereals. These include in situ hybridization of single-copy probes (Chen & Gustafson, 1995) and the analysis of deletion lines defined by C-banding (Gill et al., 1993; Hohmann et al., 1994; 1995). Potentially, the Lp/Fp introgression mapping system has important advantages over these methods. First, use of GISH is significantly less technically demanding than the detection of single copy sequences. Secondly, large numbers of lines carrying Fp chromosome segments can be made for any specific arm simply by backcrossing to Lp. This is much easier than the isolation and characterization of deletion lines. Thus we envisage that the Lp/Fp recombinants will have a major impact on aligning physical and genetic maps.
A consequence of developing the Lp/Fp recombinant chromosome series is that many lines of Lp will be produced with a single small Fp chromosome segment carrying an important gene. We have already achieved the transfer of genes for delayed senescence (Thomas et al., 1994), drought (Humphreys & Pasakinskiene, 1996) and cold tolerance, crown rust resistance, nutritive quality, late heading, perenniality, various morphological characters and some isozymes from the Festuceae into Lolium (unpubl. data). The commercial value to the UK market alone of the gene transfer for delayed senescence and drought tolerance, respectively, has been estimated at £15 and £45 millions per annum.
References
Chen, J. M. and Gustafson, J. P. (1995). Physical mapping of restriction fragment length polymorphisms (RFLPs) in homoeologous group 7 chromosomes of wheat by in situ hybridization. Heredity, 75: 225–233.
Dellaporta, S. L., Wood, J. and Hicks, J. B. (1983). A plant DNA minipreparation: Version 11. Plant Mol Biol Rep, 1: 19–21.
Gale, M. D. and Miller, T. E. (1987). The introduction of genetic variation into wheat. In: Lupton, F. G. H. (ed.) Wheat Breeding. Its Scientific Basis, pp. 173–210. Chapman and Hall, London and New York.
Ganal, M. W., Young, N. D. and Tanksley, S. D. (1989). Pulsed field gel electrophoresis and physical mapping of large DNA fragments in the TM-2a region of chromosome 9 in tomato. Mol Gen Genet, 215: 395–400.
Gill, K. S., Gill, B. S. and Endo, T. R. (1993). A chromosome region-specific mapping strategy reveals gene-rich telomeric ends in wheat. Chromosoma, 102: 374–381.
Hohmann, U., Endo, T. R., Gill, K. S. and Gill, B. S. (1994). Comparison of genetic and physical maps of group 7 chromosomes from Triticum aestivum. Mol Gen Genet, 245: 644–653.
Hohmann, U., Graner, A., Endo, T. R., Gill, B. S. and Herrmann, R. G. (1995). Comparison of wheat physical maps with barley linkage maps for group 7 chromosomes. Theor Appl Genet, 91: 618–626.
Humphreys, M. W. and Pasakinskiene, I. (1996). Chromosome painting to locate genes for drought resistance transferred from Festuca arundinacea into Lolium multiflorum. Heredity, 77: 530–534.
Jauhar, P. P. (1975). Chromosome relationships between Lolium and Festuca (Gramineae). Chromosoma, 52: 103–121.
King, I. P., Morgan, W. G., Harper, J. A., Meredith, M. R., Jones, R. N., Armstead, I. P. et al. (1997). Introgression Mapping: A Tool to Integrate Physical and Genetic Maps. Plant and Animal Genome V, San Diego, CA.
Law, C. N., Snape, J. W. and Worland, A. J. (1987). Aneuploidy in wheat and its uses in genetic analysis. In: Lupton, F. G. H. (ed.) Wheat Breeding. Its Scientific Basis, pp. 71–127. Chapman and Hall, London and New York.
Lewis, E. J. (1966). The production and manipulation of new breeding material in Lolium-Festuca. In: Hill, A. G. G. (ed.) Proc X Int Grassland Congr, Valtioneuvoston Kirjapaino, Helsinki, pp. 688–693.
Miller, T. E., Reader, S. M., Purdie, K. A. and King, I. P. (1994). Determination of the frequency of wheat-rye pairing in wheat×rye hybrids with and without chromosome 5B. Theor Appl Genet, 89: 255–258.
Pessino, S. C., Ortiz, J. P. A., Leblanc, O., Dovale, C. B., Evans, C. and Hayward, M. D. (1997). Identification of a maize linkage group related to apomixis in Brachiaria. Theor Appl Genet, 94: 439–444.
Stammers, M., Harris, J., Evans, G. M., Hayward, M. D. and Andforster, J. W. (1995). Use of random PCR (RAPD) technology to analyse phylogenetic relationships in the Lolium/Festuca complex. Heredity, 74: 19–27.
Thomas, H. M., Morgan, W. G., Meredith, M. R., Humphreys, M. W., Thomas, H. and Leggett, J. M. (1994). Identification of parental and recombined chromosomes in hybrid derivatives of Lolium multiflorum×Festuca pratensis by genomic in situ hybridization. Theor Appl Genet, 88: 909–913.
Thomas, H. M., Harper, J. A., Meredith, M. R., Morgan, W. G., Thomas, I. D., Timms, E. and King, I. P. (1996). Comparison of ribosomal DNA sites in Lolium species by fluorescence in situ hybridization. Chromosome Res, 4: 486–490.
Thomas, H. M., Harper, J. A., Meredith, M. R., Morgan, W. G. and King, I. P. (1997). Physical mapping of ribosomal DNA sites in Festuca arundinacea and related species by in situ hybridization. Genome, 40: 406–410.
Vos, P., Hogers, R., Bleeker, M., Reijans, M., Van De Lee, T., Hornes, M. et al. (1995). AFLP: a new technique for DNA fingerprinting. Nucl Acids Res, 23: 4407–4414.
Acknowledgements
I. P. Armstead and A. Bollard are funded by the Biotechnology and Biological Sciences Research Council's Plant and Animal Genome Analysis initiative.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
King, I., Morgan, W., Armstead, I. et al. Introgression mapping in the grasses. I. Introgression of Festuca pratensis chromosomes and chromosome segments into Lolium perenne. Heredity 81, 462–467 (1998). https://doi.org/10.1046/j.1365-2540.1998.00437.x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1046/j.1365-2540.1998.00437.x
Keywords
This article is cited by
-
Cytogenetic and molecular genotyping in the allotetraploid Festuca pratensis × Lolium perenne hybrids
BMC Genomics (2019)
-
Genome-dependent chromosome dynamics in three successive generations of the allotetraploid Festuca pratensis × Lolium perenne hybrid
Protoplasma (2015)
-
Exploitation of interspecific diversity for monocot crop improvement
Heredity (2013)
-
Variability of ribosomal DNA sites inFestuca pratensis, Lolium perenne, and their intergeneric hybrids, revealed by FISH and GISH
Journal of Applied Genetics (2010)
-
Stability across environments of the coffee variety near infrared spectral signature
Heredity (2009)