Abstract
Self-incompatibility systems in different angiosperm families are reviewed, and the evidence that incompatibility has arisen several times is outlined. New data on the sequence polymorphism of self-incompatibility loci from two different angiosperm families are compared with results from other highly polymorphic loci, particularly MHC loci. We discuss what molecular genetic analyses of these sequences can tell us about the nature and maintenance of the polymorphism at self-incompatibility loci. We suggest that there is evidence for recombination at the Brassica self-incompatibility loci, so that it may be possible to discern regions that are particularly functionally important in the recognition reaction, even though the long-term maintenance of polymorphisms in these amino acid residues has caused the evolution of many other sequence differences between alleles.
Similar content being viewed by others
Introduction
The self-incompatibility (SI) of flowering plants is a classic of genetics and population genetics. Self pollen, and pollen carrying ‘S’-alleles in common with a potential recipient plant is recognized and prevented from fertilizing ovules. This recognition system interests cell biologists, though the mechanism is not yet well understood at the cellular level (Goring & Rothstein, 1996; Kao & McCubbin, 1996; Li et al., 1995; Rudd et al., 1996), while the impressive S-allele polymorphism has attracted the interest of population geneticists. In this article, we review recent results that are helping further our understanding of the evolution of self-incompatibility, focusing particularly on sequence data from self-incompatibility loci of Brassica species. These data are helping to illuminate evolutionary processes, but it is still difficult to make inferences about function.
In ‘gametophytic’ SI systems, pollen incompatibility types are controlled by the grains' own haploid genotypes (reviewed by Kao & McCubbin, 1996). Gametophytic inheritance with a single incompatibility (S) locus is known in Scrophulariaceae, Onagraceaeae, Papaveraceae, Solanaceae, Rosaceae, and several other flowering plant families (reviewed in Weller et al., 1995), and systems with two or more loci occur in several families, including grasses (Li et al., 1995). Single-locus ‘sporophytic’ inheritance (pollen incompatibility type determined by the genotype of the diploid plant producing the pollen) exists in Brassicaceae, Asteraceae, and several other flowering plant families (Kowyama et al., 1980; Goodwillie, 1997). Dominance/recessivity of alleles is common in both pollen and pistil in these homomorphic SI systems (Sampson, 1967). In both types of SI, the S-loci have spectacular polymorphisms, with many S-alleles in populations (e.g. Sampson, 1967; Wright, 1939). Different populations sometimes, but not always, have have strongly overlapping sets of alleles (Nou et al., 1993; O'Donnell, et al., 1993). In terms of allele numbers, the S-loci are among the most highly polymorphic loci known, similar to mammalian MHC loci (O'hUigin, 1995), or fungal incompatibility loci (May & Matzke, 1995).
Evolution and maintenance of self-incompatibility
Unlike the origin of self-incompatibility (Holsinger & Steinbachs, 1997), the maintenance of variability at the self-incompatibility loci is well understood (Wright, 1939). Rare alleles have a fertility advantage because pollen carrying such alleles will not be rejected by incompatibility reactions of recipient plants. In a large enough population, this ‘frequency-dependent’ selection favours each new incompatibility-type allele until an equilibrium is reached with equal allele frequencies. The equilibrium number of alleles will thus be high, depending on population sizes (since alleles will be lost by chance in finite populations) and mutation rates to new specificities (since frequent mutation increases allele numbers, see Wright, 1939). The fertility advantage to low-frequency alleles also means that losses by genetic drift are soon restored if there is gene flow from other populations. Alleles should thus be maintained in species for long evolutionary times (Vekemans & Slatkin, 1994).
Until recently, high allelic diversity hindered study of alleles in populations, as probes based on one cloned allele rarely detect other alleles by Southern blotting, even under low stringency. Sequence-based approaches are now permitting renewed evolutionary and population studies of the self-incompatibility polymorphism. In several self-incompatible plants, alleles have now been cloned that segregate with the incompatibility types of plants in families, and encode sequences of cosegregating pistil proteins (Anderson et al., 1989; Li et al., 1995; Nasrallah et al., 1987; Walker et al., 1996).
An important result from sequencing studies is that self-incompatibility loci have evolved from independent origins several times in flowering plants. The gene family to which the S-locus belongs has now been identified in several angiosperm families. The pistil S-proteins in Solanaceae, Rosaceae and Scrophulariaceae are all related to RNases, though this may be due either to independent evolution or common origin (Sassa et al., 1996). A quite different gene product is implicated in Papaver rhoeas (Rudd et al., 1996) and the only pollen-expressed gene yet characterized (in the grass Phalaris coerulescens) shows thioredoxin activity (Li et al., 1995).
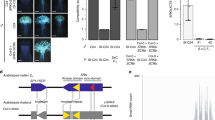
The system in the Brassicaceae, the only sporophytic system characterized at the molecular level, is quite different (Fig. 1a). It is unique in that two linked loci, SLG and SRK (encoding, a stigmatically expressed S-locus glycoprotein and a receptor kinase, respectively), probably play essential roles in incompatibility. The intronless SLG locus encodes an extracellular protein probably involved in recognition, consistent with the finding that it is highly polymorphic. The evidence for its necessary role in self-incompatibility is not yet complete. The polymorphism of the closely linked SRK gene could conceivably produce polymorphism at SLG (see below), so direct evidence of SLG being essential for functional incompatibility is needed. To date, the best evidence comes from B. campestris rendered self-compatible by mutation at an unlinked locus that affects stigma but not pollen function; the mutants have lowered SLG RNA and protein products (and of some other loci in the gene family), but SRK expression is unaffected (Nasrallah et al., 1992).
(a) Schematic diagram of the general features of the S-gene region in Brassica. Introns in the kinase domain of the SRK gene are not to scale. Numbers of genes in the S-locus region differ between species and some may be pseudogenes (Suzuki et al., 1997). The directions of transcription are for B. napus (Yu et al. 1996). In B. oleracea, transcripts in both directions are generated from SRK (reviewed in Pastuglia et al., 1997b). (b) Comparisons of synonymous and non synonymous differences per site between SLG and the S-domain of SRK-alleles, within haplotypes and between different haplotypes. The regions of the two loci compared are indicated by the horizontal bars in the figure.
The SRK locus contains an S-domain, homologous in sequence to the SLG locus, an apparent transmembrane domain and a domain with six introns that has homology to members of a serine/threonine kinase gene family (Stein et al., 1991) present in other plant species (Dwyer et al., 1994). The kinase is surmised to be an extracellular receptor (Stein et al., 1991). It also resembles an immunoglobulin-like repeat sequence (Glavin et al., 1994) and may be related to plant proteins involved in defence against pathogens, another plant recognition system (Pastuglia et al., 1997a). Mutational loss of the kinase function causes self-compatibility, so this gene product is essential for incompatibility (Goring & Rothstein, 1996), though not necessarily for the recognition function.
These findings strongly suggest that SI systems evolved independently in different families. This is consistent with information on the distribution of SI (Weller et al., 1995), but not with the view that incompatibility evolved in the ancestor of all angiosperms and was subsequently modified into gametophytic and sporophytic systems (Whitehouse, 1950; Beach & Kress, 1980).
Quantitative measures of the self-incompatibility locus polymorphism
The first sequence studies of small numbers of alleles laboriously and independently cloned from cultivated or laboratory strains of plants immediately revealed astonishing divergence between alleles for different incompatibility types, in both Solanaceae (Anderson et al., 1989) and Brassicaceae (Nasrallah et al., 1987). Both silent differences and multiple amino acid differences were found between S alleles. Relatively conserved regions were identified that can be used to design primers for PCR-based analysis of alleles. Combining PCR with restriction enzyme digestion has made it possible to obtain allele-specific bands, for typing plants of self-incompatible horticultural species (Brace et al., 1993; Janssens et al., 1995). This opens the way for sequencing and thus study of allelic diversity among plants in natural populations, even when, as is usual, they are heterozygotes.
Study of alleles from natural populations is needed in order to go beyond counting alleles, to describe quantitatively the amino acid and base sequence diversity between S-alleles, and to subject the polymorphisms to molecular evolutionary analyses that can test for recombination and selection at sites in these loci. Ideally, the incompatibility types of the alleles sequenced should be known, and the alleles should be randomly picked. No study has yet achieved this ideal, and so far, apart from the very interesting finding of silent sequence differences between P. rhoeas alleles of the same SI type (Walker et al., 1996), sequences have been compared only between S-alleles for different incompatibility types. These will on average differ more than randomly picked alleles. The first study of alleles from a natural population was done in a gametophytic system, in two species of Solanaceae, though the incompatibility types of the alleles sequenced are not yet known (Richman et al., 1996). Another large set of sequence data, from Brassica alleles of known incompatibility type, is now available from GenBank (Kusaba et al., 1997). As they are from cultivated strains, these sequences probably include only part of the natural diversity. We here review some of the results from analyses of SI sequences, focusing mainly on Brassica.
Measuring the polymorphism by the diversity per nucleotide site (Nei, 1987), differences between Brassica S-alleles are lower than in the natural populations of Solanaceae surveyed (Fig. 2) or the two P. rhoeas alleles compared (Walker et al., 1996), and similar to differences at MHC loci (Hughes & Nei, 1989). In all species so far studied, polymorphism is found throughout the S-locus sequences, particularly in certain (‘hypervariable’ or HV) regions, where diversity is spectacularly high (Table 1).
Distribution of pairwise diversity values between alleles within species, in S- and MHC-loci. (a) shows alleles in a populations with gametophytic SI, together with some MHC alleles. The data for sporophytic SI (b) are based on 40 GenBank-derived SLG alleles of known incompatibility types in Brassica (B. oleracea 21 alleles, B. campestris, 19 alleles). The sequences analysed include only coding regions of functional alleles, and only type 1 (dominant) incompatibility alleles, as most available sequences are from this type. The few type 2 alleles appear extremely different in sequence (Kusaba et al., 1997). After alignment, diversities were estimated separately for each codon, using pairs of alleles from within each species, or pairs consisting of one from each species (between-species values).
Age of the alleles
Alleles at the S-loci must be very ancient, because long periods of time, and low rates of recombinational exchange (Nr) are required for silent substitutions to accumulate between alleles within each species (Hudson, 1990). Furthermore, unlike most loci, whose alleles cluster together within species, S-allele sequences are sometimes more similar to alleles from related species than to others from their own species: Solanum alleles mingle in the allele tree with those from species in the genera Petunia, Lycopersicon, and Nicotiana, and the same is seen in Brassica species (Hinata et al., 1995; Kusaba et al., 1997). This ‘trans-specific clustering’ suggests that the S-alleles have been polymorphic since before the species became isolated from one another (Dwyer et al., 1991; Ioerger et al., 1990; Richman et al., 1996).
Quantitative analysis of Brassica oleracea and campestris S-allele sequences reveals, astonishingly, that average differences between SLG type 1 (dominant) alleles are as large when alleles are compared within species as when alleles from different species are compared (Fig. 2). This is true even for synonymous sites, and even in the most conserved regions, and also for the S-domain of SRK, discussed in more detail below (Table 1) Unless sequences can be exchanged by hybridization, these data suggest that the polymorphism must date from before the species split, though, unlike MHC alleles, no motifs common across different species (see O'huigin, 1995) are discernible in S-allele sequences. Turnover of alleles is also probably infrequent, since replacement of alleles by descendant alleles with new specificities will increase within-species similarity.
The alternative that sequence similarity between alleles from different species could occur by chance ‘convergence’ seems unlikely for the numerous amino acid and silent differences seen between S-alleles, but no general test for convergence yet exists to rule it out. It may occur in MHC loci, as different codons are used for the same amino acids in different species (Gustafsson & Andersson, 1994; O'huigin, 1995).
Nature of the differences between SLG alleles
The great age of the alleles makes it difficult to test for differences in functional constraints between different parts of the S-loci, and it is not yet certain that a meaningful pattern of diversity differences exists. Stochastic variation in diversity is expected at different sites within a sequence, even when all are evolving neutrally (Hudson, 1990). This seems unlikely to explain the diversity differences in the S-locus (hypervariable regions), which appear robust as sequence data have accumulated from more and more alleles (Hinata et al., 1995). It is, however, very unlikely that all the variants in these regions are maintained by balancing selection. Much of the diversity may be a consequence of linkage to a few sites under balancing selection. If there is little or no recombinational exchange between alleles, each functional allele class will behave like an isolated population. Because many SI types are maintained in species, effective population sizes of individual classes will be low. Even nonsynonymous mutations may therefore drift to high frequency or fixation within classes, so differences additional to those involved in determining SI type will accumulate, provided they are not strongly detrimental to function. This will produce variability at sites not themselves under such selection, as observed for the Drosophila Adh locus where, an extreme peak in diversity exists around the site of the F/S allozyme polymorphism (Hudson, 1990). At the S-loci, because multiple different alleles are maintained, the polymorphism must involve many sites, so we might expect a wide region of increased diversity due to drift.
As in other attempts to determine functionality from sequence data, it is remarkably difficult to decide between the extreme opposites: polymorphic sites such as the HV regions may be the most important (where amino acid differences determine the incompatibility types), or they may be regions of minor functional importance that are more free to vary than other parts of the locus. This dashes hopes that regions of the S-locus important in determining functional differences between incompatibility types can be pinpointed simply by identifying the most variable parts of the sequence. It even implies that the high polymorphism of the SLG locus is not a sure sign of its involvement in recognition. More direct evidence is necessary. For instance, comparisons between alleles of the same SI type should help identify sites which can change without affecting incompatibility type (the one such study to date found only silent differences Walker et al., 1996). The direct evidence that two B. campestris SLG alleles with different specificities have identical hypervariable region amino acid sequences suggests that other regions can determine specificity (Kusaba et al., 1997). Despite many difficulties, some evidence from transgenic approaches is becoming available. Replacement of the HV region of one Solanum allele by that from another allele with a very similar sequence has been shown to change the pistil incompatibility appropriately (Matton et al., 1997).
Evidence for diversifying selection
Instead of merely showing that diversity is high, a more sensitive method of testing for the operation of diversifying selection Fig. 2.is to quantify amino acid changes, and compare their frequency with that of silent changes, by Pn/Ps ratios (Nei, 1987). Natural selection usually eliminates alleles coding for proteins with variant amino acid sequences, so high values (e.g. 2–3 for the MHC antigen recognition sites) suggest balancing selection (Hughes & Nei, 1989). For the four variable regions identified in the SLG locus (Kusaba et al., 1997), Pn/Ps averages 0.95 for the Brassica species, i.e. as expected for neutrally evolving coding sequences (Nei, 1987). For the regions classified as conserved, values are lower (about 0.44) but still very high compared with other genes. Values derived from comparing sequences of nuclear genes between maize and rice average about 0.12 (Wolfe et al., 1989); few data are available for plant DNA polymorphisms, but a similar value (0.12) was estimated for Adh loci within species of the plant genus Leavenworthia (Charlesworth et al., 1998). Correction for saturation of the synonymous substitutions would increase the denominator in the calculations and reduce the values for the S-allele data, but even the nonsynonymous sites approach saturation, so the effect of correction is slight. As explained above, however, the possibility that polymorphisms may be present because of linkage to selectively maintained sites elsewhere in the locus makes it impossible to conclude from these results that the variable regions are more functionally important than the conserved ones.
Diversifying selection should also lead to nonconservative amino acid substitutions, with changed charge or polarity, whereas other parts of the locus should have mainly amino acids with similar properties (conservative differences). Excess nonconservative amino acid substitutions suggest the operation of diversifying selection at other recognition loci (e.g. Hughes et al., 1990). But in the B. oleracea and campestris S-allele sequences, nonconservative amino acid substitutions are slightly less likely than conservative changes, though much more frequent than in the kinase domain of SRK (Table 1). These data do not therefore indicate strong diversifying selection. Rather, sites within these regions appear to be evolving neutrally.
Finally, as in several other systems, the accumulation of differences at nonsynonymous sites in both the gametophytic and Brassica S-loci seem to be slowing down with evolutionary time. This is inferred because average numbers of nonsynonymous differences between allele pairs increase more slowly than synonymous ones (which increase roughly in proportion to divergence times but slow down due to saturation of substitutions). This difference becomes more pronounced as the number of synonymous differences between the alleles compared gets bigger (Hinata et al., 1995; Uyenoyama, 1997). This is the opposite of the effect of saturation and suggests diversifying selection, as differences accumulating neutrally should show a linear relation (Tanaka & Nei, 1989). This effect might help test which regions of the locus are under diversifying selection pressure. Surprisingly, such tests applied to the Brassica data show just as clear evidence for the conserved as for the HV regions, suggesting that both are selected similarly. This could be due to a slowing down of the rate of replacement of alleles by descendents with new incompatibility types. Accumulation of nonsynonymous substitutions (see above) or slightly deleterious mutations at linked sites (Uyenoyama, 1997) implies that heterozygous progenitor/descendant genotypes will be homozygous for deleterious variants of the progenitors, disfavouring new alleles. Their rarity advantage (explained above) diminishes as allele numbers in a population increase, so it becomes increasingly unlikely that new SI alleles will replace their progenitors, resulting in a ‘frozen polymorphism’ in which only synonymous substitutions occur.
Polymorphism at other loci in Brassica
To evaluate and interpret data from S-loci, heterozygosity and genetic diversity data for non-S reference loci will be helpful, as a basis for comparison, which can help the search for evidence of selection (Hudson, 1990) and hybridization. As already mentioned, sequence diversity has scarcely been studied in plant populations. Plants probably do not differ greatly in this respect from animals such as Drosophila, whose sequence diversity is well studied (Moriyama & Powell, 1995), but outbreeding plants may have higher diversity (Henry & Damerval, 1997; Liu et al., 1998).
Comparisons of diversity between different S-locus regions may also be informative, despite the difficulties mentioned earlier. Both the SLG- and SRK-loci are clearly exceptionally variable, while other loci in the S-gene family, whose products are nonessential for incompatibility, apparently have much less allelic polymorphism, though natural populations have not been studied (Hinata et al., 1995). Current scanty data on S-locus related (SLR) genes, nonessential for recognition, suggest sequence conservation within and between species (Hinata et al., 1995). For the unlinked SRA and SRB loci, silent differences (i.e. coalescence times) are greater between than within species, unlike SLG and SRK, though SRB shows moderate polymorphism. The S-linked anther-expressed B. oleracea locus SLA is nonessential for incompatibility and has low polymorphism, as have several other loci in the S-gene region whose expression is not confined to pistils, implying that they are not involved in pollen recognition processes (Nasrallah et al., 1991; Pastuglia et al., 1997b). These polymorphism differences, even involving loci linked to the S-locus, suggest some independence in their evolution.
This independence can also be detected in comparisons of SLG and SRK. The polymorphism in the S-domain of the few SRK alleles currently available appears similar to that at the SLG locus (both have mean diversity per base of 0.13). Pn/Ps values are as high as those for corresponding SLG regions. Over most of the variable regions, radical amino acid substitutions per site are again nearly as likely as conservative ones. When alleles from the two species are compared, Ps values are again no larger than for within-species comparisons, except perhaps for the conserved regions. These extensive differences suggest that the SRK S-domain may have a balanced polymorphism similar to that in SLG, and thus that it participates in recognition of incompatibility types.
In contrast, the SRK kinase coding sequence has fewer nonsynonymous differences (Table 1). Pn/Ps averages 0.48, similar to the conserved regions of SLG or the SRK S-domain. Consistent with this domain's overall lower variability, probabilities of radical amino acid differences between alleles within both species average less than half those of conservative differences. Finally, unlike the observations on SLG or the S-domain of SRK, Ps estimates from the kinase domain are somewhat less within each species than between them (Table 1). There are thus clear signs of selective constraint acting at the SRK locus, and its polymorphism may be less ancient than in the other regions, though still very old, since the introns are highly variable, implying long times to common ancestry of alleles (Nishio et al., 1997). This regional variability suggests that the kinase domain polymorphism is caused by linkage to sites elsewhere in SRK that are under balancing selection, with some recombination in the locus. With no recombination, all parts of the locus have identical coalescence times, so we expect no regional heterogeneity in silent diversity, even if selective constraints differ.
Linkage disequilibrium and recombination in the SLG-SRK region
The sequences of the SLG gene and the S-domain of the SRK gene from any given haplotype tend to be similar, whereas those from different haplotypes differ extensively. This has suggested that functional self-incompatibility requires alleles from the two loci to be similar and that recombination between the two loci is suppressed to maintain this matching (Stein et al., 1991). High sequence divergence in the region between the loci (Boyes et al., 1997) and near Petunia inflata S-loci (Coleman & Kao, 1992) support rarity of recombinational exchange, but divergence could be due to relaxed selection in these flanking regions. Sequence differences are not only a feature of S-loci, but are also found in maize intergenic regions (Sanmiguel et al., 1997). Further sequence data from Brassica show that SLG alleles are not always similar to SRK alleles from the same haplotype (Goring & Rothstein, 1996; Kusaba et al., 1997). For the few haplotypes sequenced for both loci, both synonymous and nonsynonymous within-species differences between haplotypes average almost double their within-haplotype values (Fig. 1B), in other words there is some linkage disequilibrium. This is consistent with the idea of selectively maintained within-haplotype similarity between the two loci. But an alternative is that sequence similarity is caused by exchange of sequence information between them (e.g. by gene conversion)
We need more evidence on whether genetic exchanges, by recombination or gene conversion, occur within the S-locus region. No estimates of recombination rates from the sequence data (see Hudson, 1990) have yet been attempted. Such estimates may be impossible, because the high variability may obscure patterns, and apparent ‘recombinant’ sequence motifs may be caused by independent origination of similar sequences. Some evidence, however, suggests recombination. If recombination were totally suppressed, we should expect strong linkage disequilbrium between sites in the S-loci, which is not apparent in the data. Rather, the data show a pattern similar to that in other loci (e.g. Miyashita et al., 1993): linkage disequilibrium is found only for very close sites, suggesting that, even within the SLG locus, more distant sites recombine over evolutionary time, consistent with the evidence for some measure of independence in the patterns of evolution of the SRK and SLG loci (above). This view is also consistent with the observation that different sequence regions within the S-locus produce different allele trees (Kusaba et al., 1997), though the significance of such differences is uncertain as the sequence regions are short. The presence of nonpolymorphic loci in the S-locus region also suggests recombination, such that each locus evolves essentially independently.
The pollen locus
A major question concerning the recognition function of self-incompatibility systems is whether there are separate pistil and pollen loci, as often assumed (e.g. Stephenson et al., 1997). This view involves several difficulties. It requires strong linkage disequilibrium, and very infrequent recombination, so that each pistil type rejects only its own pollen. Furthermore, recognition is relatively easy to imagine if one locus is involved and identity causes rejection. But if two loci are involved each pistil allele requires a different recognition reaction for pollen carrying just one allele product of the other locus. Finally, mutation at either locus will not create new incompatibility types but merely produce self-compatibility, so it is difficult to imagine how new SI types can arise.
To date, no candidate for the pollen locus has been discovered in any self-incompatibility system other than Phalaris coerulescens, but this gene is not expressed in pistils and the gene controlling the pistil incompatibility reaction is not known in this, or any other, grass species (Li et al., 1995). Loci for the pollen and pistil recognition functions must be tightly linked, otherwise recombinants should exist with different pollen and pistil incompatibility types; these are known in distylous species (Ernst, 1936) but have not been found in species with homomorphic SI (Lewis, 1963; Nasrallah et al., 1991; Nasrallah et al., 1992).
If the S-locus region rarely recombines, it may have accumulated repetitive sequences, making it hard to find coding sequences within it by approaches such as chromosome walking. Transgenic approaches may also fail due to cosuppression (e.g. Jorgensen, 1990), as expression of additional S-locus copies in pollen (e.g. in the diploid pollen of tetraploids) often causes self-compatibility (Lewis, 1963). It might be possible to identify the pollen locus by in vitro testing of pollen proteins, though work in Brassica has so far not definitively identified an S-linked component causing allele-specific rejection (Stephenson et al., 1997). Alternatively, molecular analysis of self-compatible mutants in which either the pollen or pistil SI type is not expressed, while the other function remains intact (see Lewis, 1963; Nasrallah et al., 1991 should be informative. If ‘pollen-part mutations’ involve the S-locus encoding the pistil S-protein, this would cast doubt on the existence of a separate locus. Such information is difficult to obtain, because mutations of known progenitor alleles are needed. Otherwise the high polymorphism in these loci makes it impossible to know that any sequence difference found really causes the mutant phenotype (Pastuglia et al., 1997b). In the one study yet published, a mutant Japanese pear with deleted stylar S-RNase, appears to have normal pollen reactions (Sassa et al., 1997); unless the plant is merely a chimaera, this implies a separate pollen locus, despite the difficulties of this hypothesis. Given the long history of astonishing results from SI systems, outcomes surprising to geneticists are now almost expected.
References
Anderson, M. A., McFadden, G. I., Bernatzky, R., Atkinson, A., Orpin, T., Dedman, H., Tregear, G., Fernley, R. and Clarke, A. E. (1989). Sequence variability of three alleles of the self-incompatibility gene of Nicotiana alata. Pl Cell, 1: 483–491.
Beach, J. H. and Kress, W. J. (1980). Sporophytic vs. gametophytic: a note on the origin of self-incompatibility in the flowering plants. Syst Bot, 5: 1–5.
Boyes, D. C., Nasrallah, M. E., Vrebalov, J. and Nasrallah, J. B. (1997). The self-incompatibility (S) haplotypes of Brassica contain highly divergent and rearranged sequences of ancient origin. Pl Cell, 9: 237–247.
Brace, J., Ockendon, D. J. and King, G. J. (1993). Development of a method for the identification of S alleles in Brassica oleracea based on digestion of PCR amplified DNA with restriction endonucleases. Sex Pl Repr, 6: 133–138.
Charlesworth, D., Liu, F. -L. and Zhang, L. (1998). The evolution of the alcohol dehydrogenase gene family in plants of the genus Leavenworthia (Brassicaceae): loss of introns, and an intronless gene. Mol Biol Evol. in press.
Coleman, C. A. and Kao, T. -H. (1992). The flanking regions of Petunia inflata S alleles are heterogeneous and contain repetitive sequences. Pl Molec Biol, 18: 725–737.
Dwyer, K. G., Balent, M. A., Nasrallah, J. B. and Nasrallah, M. E. (1991). DNA sequences of self-incompatibility genes from Brassica campestris and B. oleracea: polymorphism predating speciation. Pl Molec Biol, 16: 481–486.
Dwyer, K. G., Kandasamy, M. K., Mahosky, D. I., Axxiai, J., Kudish, B. I., Miller, J. E., Nasrallah, M. E. and Nasrallah, J. B. (1994). A superfamily of S locus-related sequences in Arabidopsis: diverse structures and expression patterns. Plant Cell, 6: 1829–1843.
Ernst, A. (1936). Weitere untersuchungen zur phänanalyse zum fertilitätsproblem und zur genetik heterostyler primeln. II. Primula hortensis Arch Julius-Klaus Stift Vererbungsforsch, Sozial-Anthropol Rassenhygiene, 11: 1–280.
Glavin, T. L., Goring, D. R., Schafer, U. and Rothstein, S. J. (1994). Features of the extracellular domain of the S-locus receptor kinase from Brassica. Mol Gen Genet, 244: 630–637.
Goodwillie, C. (1997). The genetic control. of self-incompatibility in Linanthus parviflorus (Polemoniaceae). Heredity, 79: 424–432.
Goring, D. F. and Rothstein, S. J. (1996). S-locus receptor kinase genes and self-incompatibility in Brassica napus ssp. oleifera. In: D. P. S. Verma (ed.) Signal Transduction in Plant Growth and Development, pp. 217–230. Springer, New York.
Gustafsson, K. and Andersson, L. (1994). Structure and evolution of horse MHC class II DRB genes: convergent evolution in the antigen-binding site. Immunogenetics, 39: 355–358.
Henry, A. -M. and Damerval, C. (1997). High rates of polymorphism and recombination at the Opaque-2 locus in maize. Mol Gen Genet, 256: 147–157.
Hinata, K., Watanabe, M., Yamakawa, S., Satta, Y. and Isogai, A. (1995). Evolutionary aspects of the S-related genes of the Brassica self-incompatibility system: synonymous and nonsynonymous base substitutions. Genetics, 140: 1099–1104.
Holsinger, K. E. and Steinbachs, J. E. (1997). Mating systems and evolution in flowering plants. In: K. Iwatsuki and P. Raven (eds). Evolution and Diversification of Land Plants, pp. 223–248. Springer, Tokyo.
Hudson, R. R. (1990). Gene genealogies and the coalescent process. Oxf Surv Evol Biol, 7: 1–45.
Hughes, A. and Nei, M. (1989). Nucleotide substitution at major histocompatibility complex class II loci: evidence for overdominant selection. Proc Natl Acad Sci USA, 86: 958–962.
Hughes, A., Ota, T. and Nei, M. (1990). Positive Darwinian selection promotes charge profile diversity in the antigen-binding cleft of class I major-histocompatibility-complex molecules. Mol Bio Evol, 76: 515–524.
Ioerger, T. R., Clark, A. G. and Kao, T. -H. (1990). Polymorphism at the self-incompatibility locus in Solanaceae predates speciation. Proc Natl Acad Sci USA, 87: 9732–9735.
Janssens, G. A., Goderis, I. J., Broekaert, W. F. and Broothaerts, W. (1995). A molecular method for S-allele identification in apple based on allele-specific PCR. Theor Appl Genet, 91: 691–698.
Jorgensen, R. (1990). Altered gene expression in plants due to trans interactions between homologous genes. Trends Biotechnol, 8: 340–344.
Kao, T. -H. and McCubbin, A. G. (1996). How flowering plants discriminate between self and non-self pollen to prevent inbreeding. Proc Natl Acad Sci USA, 93: 12059–12065
Kowyama, Y., Shimano, N. and Kawasi, T. (1980). Genetic analysis of incompatibility in the diploid species Ipomoea closely related to the sweet potato. Theor Appl Genet, 58: 149–155.
Kumar, S. Tamura, K. and Nei, M. (1994). MEGA - molecular evolutionary genetics analysis software for microcomputers. Comput Appl Biosci, 10: 189–191.
Kusaba, M., Nishio, T., Satta, Y., Hinata, K. and Ockendon, D. (1997). Striking similarity in inter- and intra-specific comparisons of class I SLG alleles from Brassica oleracea and Brassica campestris: Implications for the evolution and recognition mechanism. Proc Nat Acad Sci USA, 94: 7673–7678.
Lewis, D. (1963). A protein dimer hypothesis on incompatibility. In: S. J. Geerts (ed.) Genetics Today, pp. 656–663 Pergamon Press, Oxford.
Li, X., Nield, J., Hayman, D. and Langridge, P. (1995). Thioredoxin activity in the C terminus of Phalaris S protein. Plant J, 8: 133–138.
Liu, F., Zhang, L. and Charlesworth, D. (1998). Genetic diversity in Leavenworthia populations with different inbreeding levels. Proc R Soc B, 265: 293–301.
Matton, D. P., Maes, O., Laublin, G., Xike, Q., Bertrand, C., Morse, D. and Cappadocia, M. (1997). Hypervariable domains of self-incompatibility RNases mediate allele-specific pollen recognition. Pl Cell, 9: 1757–1766.
May, G. and Matzke, E. (1995). Recombination and variation at the a mating-type of coprinus-cinereus. Mol Biol Evol, 12: 794–802.
Miyashita, N. T., Aguade, M. and Langley, C. H. (1993). Linkage disequilibrium in the white locus region of Drosophila melanogaster. Genet Res, 62: 101–109.
Moriyama, E. N. and Powell, J. R. (1995). Intraspecific nuclear DNA variation in Drosophila. Mol Biol Evol, 13: 261–277.
Nasrallah, M. E., Kandasamy, M. K. and Nasrallah, J. B. (1992). A genetically defined trans-acting locus regulates S-locus function in Brassica. Plant J, 2: 497–506.
Nasrallah, J. B., Kao, T. -H., Chen, C. -H., Goldberg, M. and Nasrallah, M. E. (1987). Amino-acid sequence of glycoproteins encoded by three alleles of the S locus of Brassica oleracea. Nature, 326: 617–619.
Nasrallah, J. B., Nishio, T. and Nasrallah, M. E. (1991). The self-incompatibility genes of Brassica: expression and use in genetic ablation of floral tissues. Ann Rev Plant Physiol Plant Mol Biol, 42: 393–422.
Nei, M. (1987). Molecular Evolutionary Genetics . Columbia University Press, New York.
Nishio, T., Kusaba, M., Sakamoto, K. and Ockendon, D. (1997). Polymorphism of the kinase domain of the S-locus receptor kinase gene (SRK) in Brassica oleracea L. Theor Appl Genet, 95: 335–342.
Nou, I. S., Watanabe, M., Isogai, A. and Hinata, K. (1993). Comparison of S-alleles and S-glycoproteins between two populations of Brassica campestris in Turkey and Japan. Sex Pl Repr, 6: 79–86.
O'huigin, C. (1995). Quantifying the degree of convergence in primate Mhc-DRB genes. Immunol Rev, 143: 123–140.
O'donnell, S., Lane, M. D. and Lawrence, M. J. (1993). The population genetics of the self-incompatibility polymorphism in Papaver rhoeasVI. Estimation of the overlap between the allelic complements of a pair of populations. Heredity, 71: 591–595.
Pastuglia, M., Roby, D., Dumas, C. and Cock, J. M. (1997a). Rapid induction by wounding and bacterial infection of an S gene family receptor-like kinase gene in Brassica oleracea. Pl Cell, 9: 49–60.
Pastuglia, M., Ruffio-Châble, V., Delorme, V., Gaude, T., Dumas, C. and Cock, J. M. (1997b). A functional S locus anther gene is not required for the self-incompatibility response in Brassica oleracea. Pl Cell, 9: 2065–2076.
Richman, A. D., Uyenoyama, M. K. and Kohn, J. R. (1996). Allelic diversity and gene genealogy at the self-incompatibility locus in the Solanaceae. Science, 273: 1212–1216.
Rudd, J. J., Franklin, F. C. H., Lord, J. M. and Franklin-Tong, V. E. (1996). Increased phosphorylation of a 26-kD pollen protein is induced by the self-incompatibility response in Papaver rhoeas. Pl Cell, 8: 713–724.
Sampson, D. R. (1967). Frequency and distribution of self-incompatibility alleles in Raphanus raphanistrum. Genetics, 56: 241–251.
Sanmiguel, P., Tikhonov, A., Jin, Y. K., Motchoulskaia, N., Zakharov, D., Melakeberhan, A. et al. (1997). Nested retrotransposons in the intergenic regions of the maize genome. Science, 274: 765–768.
Sassa, H., Hirano, H. and Koba, T. (1997). Style-specific self-compatible mutation caused by deletion of the S-RNase gene in Japanese pear (Pyrus serotina). Plant J, 12: 223–227.
Sassa, H., Nishio, T., Kowyama, Y., Hirano, H., Koba, T. and Ikehashi, H. (1996). Self-incompatibility (S) alleles of the Rosaceae encode members of a distinct class of the T2/S ribonuclease superfamily. Mol Gen Genet, 250: 547–557.
Stein, J., Howlett, B., Boyes, D. C., Nasrallah, M. E. and Nasrallah, J. B. (1991). Molecular cloning of a putative receptor protein kinase gene encoded at the self-incompatibility locus of Brassica oleracea. Proc Natl Acad Sci USA, 88: 8816–8820.
Stephenson, A. G., Doughty, J., Dixon, S., Ellrman, C., Hiscock, S. and Dickinson, H. G. (1997). The male determinant of self-incompatibility in Brassica is located in the pollen coating. Plant J, 12: 1351–1359.
Suzuki, G., Watanabe, M., Kai, N., Matsuda, N., Toriyama, K., Takayama, S., Isogai, A. and Hinata, K. (1997). Three members of the S multigene family are linked to the locus of Brassica. Mol Gen Genet, 256: 257–264.
Tanaka, T. and Nei, M. (1989). Positive Darwinian selection observed at the variable region genes of immunoglobulins. Mol Biol Evol, 6: 447–459.
Uyenoyama, M. K. (1997). Genealogical structure among alleles regulating self-incompatibility in natural populations of flowering plants of self-incompatibility. Genetics, 147: 1389–1400.
Vekemans, X. and Slatkin, M. (1994). Gene and allelic genealogies at a gametophytic self-incompatibility locus. Genetics, 137: 1157–1165.
Walker, E. A., Ride, J. P., Kurup, S., Franklin-Tong, V. E., Lawrence, M. J. and Franklin, F. C. H. (1996). Molecular analysis of two functional homologues of the S3 allele of the Papaver rhoeas self-incompatibility gene isolated from different populations. Plant Mol Biol, 30: 983–994.
Weller, S. G., Donoghue, M. J. and Charlesworth, D. (1995). The evolution of self-incompatibility in the flowering plants: a phylogenetic approach. In: P. C. Hoch and A. G. Stephenson (eds). Experimental and Molecular Approaches to Plant Biosystematics. 355–382. Missouri Botanic Garden, St. Louis, MO.
Whitehouse, H. L. K. (1950). Multiple-allelomorph incompatibility of pollen and style in the evolution of the angiosperms. Ann Bot, 14: 198–216.
Wolfe, K. H. L. I. W.-H. and Sharp, P. M. (1989). Rates of synonymous substitution in plant nuclear genes. J Mol Evol, 28: 208–211.
Wright, S. (1939). The distribution of self-sterility alleles in populations. Genetics, 24: 538–552.
Yu, K., Glavin, T. L., Goring, D. F. and Rothstein, S. J. (1996). Molecular characterization of the S locus in two self-incompatible Brassica napus lines. Pl Cell, 8: 2369–2380.
Acknowledgements
We thank Mikkel H. Schierup and Brian Charlesworth for comments on this manuscript, David H. Guttman for analysis of diversity levels of gametophytic SI alleles, and T. Ota for sending us the program to analyse radical and conservative amino acid differences.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Charlesworth, D., Awadalla, P. Flowering plant self-incompatibility: the molecular population genetics of Brassica S-loci. Heredity 81, 1–9 (1998). https://doi.org/10.1038/sj.hdy.6884000
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6884000
Keywords
This article is cited by
-
A genome-wide scan for genes under balancing selection in Drosophila melanogaster
BMC Evolutionary Biology (2017)
-
Large-scale genotyping of highly polymorphic loci by next-generation sequencing: how to overcome the challenges to reliably genotype individuals?
Heredity (2015)
-
Experimental evidence of inbreeding avoidance in the hermaphroditic snail Physa acuta
Evolutionary Ecology (2006)