Abstract
RAPD-PCR polymorphisms were used to examine breeding structure in three species of snow pool Aedes mosquitoes across three river drainages in northern Colorado. Larvae were collected from four snow pools for Aedes cataphylla Dyar, seven pools for A. hexodontus Dyar and six pools for A. pullatus (Coquillet). Polymorphisms were scored at 47–48 RAPD loci in each species. To test for isolation by distance, FST/(1−FST) values between pairs of pools were plotted against geographical distances and subjected to a Mantel test with 1000 random permutations. FST values were independent of geographical distances in A. cataphylla and A. hexodontus but were somewhat correlated in A. pullatus suggesting isolation by distance in this species. A cluster analysis was performed on pair-wise FST values among pools including seven pools that were sampled in both 1994 and 1995. Bootstrap analysis indicated that pools clustered across drainages and generally independently of geographical proximity. However, there was consistent support for clustering of larvae collected from the same pool across years in A. cataphylla and in high altitude collections of A. pullatus. Mountains do not appear to act as major barriers to gene flow in any of these species. Instead, seasonal differences in adult emergence may serve as barriers to migration among A. pullatus and A. cataphylla populations. Larvae of A. hexodontus are distributed continuously in grassy pools along the banks of rivers and genetic drift probably occurs through random larval mortality when these pools are washed out during spring run-off.
Similar content being viewed by others
Introduction
Boreal or snow pool Aedes mosquitoes possess a larval stage that hatches and matures in temporary ponds produced by melting snow during the spring. These species are mostly Holarctic in distribution and over-winter as eggs. Larvae hatch in the early spring and adults emerge in late spring through early summer. Following a blood meal, females oviposit in moist recessed areas near marshes or streams generally well above the prevailing water level. Eggs enter an obligatory diapause and hatch only after experiencing several weeks of freezing temperatures (Wood et al., 1979). Hatching is stimulated by low oxygen tension that develops as bacteria become abundant in forming pools. Larval development must be completed before pools dry out and development times can vary widely for a species depending on water temperatures.
There are from 30 to 40 Aedes species world-wide that follow this life cycle (Wood et al., 1979). In the Nearctic, the species are distributed throughout Canada and the northern tier of the United States. They are also widely distributed in the Rocky Mountains of the United States and Canada and extend north through the southern half of Alaska. The exact number of species is uncertain because cryptic species have been detected. For example, the A. communis (DeGeer) complex consists of four sibling species (A. communis s.s., A. churchillensis Ellis & Brust, A. nevadensis Chapman & Barr and A. tahoensis Dyar) (Ellis & Brust, 1973; Brust & Munstermann, 1992). Schutz & Eldridge (unpublished data, cited in Hardy et al., 1993) found geographical variation in allozyme frequencies in A. tahoensis along a north–south gradient in California from the Klamath Mountains in the north to the southern Sierra Nevada Mountains, suggesting restricted gene flow between geographically adjacent populations. These findings and the broad geographical distributions for many snow pool Aedes species suggest range disjunction as a possible mode of speciation (Ross, 1964).
We hypothesized that the mountains and broad valleys of the Rocky Mountains act as barriers to gene flow among snow pool Aedes. We examined genetic variation in RAPD-PCR markers among collections of three species distributed along three river drainages in Northern Colorado to test this hypothesis. Aedes cataphylla Dyar, A. hexodontus Dyar and A. pullatus (Coquillet) were selected because of their abundance, and because they have different seasonal phenologies, larval habitats and elevational ranges (Baker, 1961; Harmston & Lawson, 1967). All three species are members of Edward's Group G (the A. communis group) of the subgenus Ochlerotatus Lynch Arribalzaga (Edwards, 1932).
Aedes cataphylla emerges from the egg in early March and most individuals have emerged as adults by the end of May. In Colorado, A. cataphylla occurs in a narrow altitudinal range between 2400 and 2800 m. Aedes hexodontus emerges from the egg in early April and most adults have emerged by the end of June. In Colorado, A. hexodontus occurs in a wide altitudinal range between 2300 and 3400 m. Harmston & Lawson (1967) record A. pullatus as the most abundant and ubiquitous mountain mosquito in Colorado and it was the most abundant mosquito in our collections with an altitudinal range between 2300 and 3400 m. The emergence time varied with altitude. At 2200–2800 m, A. pullatus emerged from the egg in late April and most individuals had emerged as adults by the end of June. At high altitudes (>3000 m) we collected larvae in early August.
Genetic variation was detected using the Random Amplified Polymorphic DNA Polymerase Chain Reaction (RAPD-PCR) (Williams et al., 1991). This method uses single 10-base primers of arbitrary sequence but with a minimum G-C content of 60% that hybridize to and amplify numerous regions of the genome. Polymorphisms revealed by RAPD-PCR are expressed among individuals as the presence or absence of a fragment of a particular size. Individuals exhibiting a band are generally assumed to be homozygous or heterozygous for a dominant allele at a RAPD locus whereas individuals not exhibiting a band are interpreted as homozygous recessives. Williams et al. (1991) observed that the majority of RAPD bands segregate as dominant markers. Mapping studies with RAPDs in the mosquito Aedes aegypti (Antolin et al. 1996) also support this pattern. Interpreting RAPD polymorphisms in this way, Apostol et al. (1996) used RAPD-PCR to estimate breeding structure among populations of A. aegypti in six cities in Puerto Rico. Here we use the same approach and analytical methods to describe breeding structure in three species of snow pool Aedes.
Materials and methods
Mosquito collection and DNA extraction
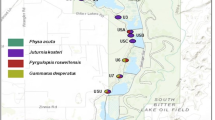
Collections were made in three distinct river drainages in northern Colorado (Fig. 1). (i) Mountains over 3500 m elevation enclose the Big Thompson River (BTR) drainage. These include the Mummy Range to the north and the continental divide to the west. (ii) The Mummy Range to the south and west separates the South Fork of the Cache la Poudre River (CLP) drainage from the BTR drainage. (iii) The North Fork of the CLP drainage is separated from the CLP South Fork by a ridge of mountains extending from the three Bald Mountains (3353 m) eastward to Lonetree Mountain (2854 m). Snow pools were located 0.1–17.2 km apart within drainages (Table 1).
Map of all collecting sites (Table 1) in the three river drainages. Collections are indicated by a filled circle. A ‘+’ indicates the location of a mountain peak >3000 m.
All mosquitoes were field-collected as larvae in plastic dippers. Each snow pool was sampled approximately once every two weeks. Larvae were reared to the fourth instar for identification (Harmston & Lawson, 1967; Darsie & Ward, 1981). We found a total of 33 pools that contained mosquito larvae; 22 of these contained A. pullatus, 24 contained A. hexodontus and 17 contained A. cataphylla. Apostol et al. (1996) showed that estimates of FST, linkage disequilibrium and genetic distance are upwardly biased by small samples and concluded that sample sizes should be >50 individuals for accurate estimates of these parameters. Only a subset of snow pools produced sufficient numbers of larvae for analysis (Table 1). One collection of A. cataphylla, seven of A. hexodontus and six of A. pullatus were made in 1994. In 1995 we analysed four pools containing A. cataphylla, three of A. hexodontus and three of A. pullatus. Exactly 50 individuals were analysed in each collection. Estimates of FST and effective migration rate (Nm) were made using 1994 collections of A. hexodontus and A. pullatus, and 1995 collections of A. cataphylla. Results from both years were combined to perform cluster analyses.
We isolated DNA from individual mosquitoes using Livak's (1984) sodium dodecyl sulphate (SDS) and salt extraction method. The DNA pellet was resuspended in 250 μL of TE (10 mM Tris, 1 mM EDTA, pH 8.0). We frequently observed epibionts such as green algae and ciliate Vorticella sp., and endoparasites such as mermithid nematodes and microsporidians infesting field-collected larvae. The short oligonucleotide primers used in RAPD-PCR anneal at low temperatures to all template DNA present in the reaction. The differences in RAPD-PCR bands among larvae could therefore reflect the presence of endo- and ectoparasites rather than true genetic differences among mosquitoes. However, newly emerging adults are relatively free of associated organisms and thus in 1994 RAPD-PCR was performed on emerging adult mosquitoes and larvae from the same collection. Some of these larvae were covered with epibionts or infected with microsporidians. No differences in RAPD patterns were detected and no bands consistently appeared in infected individuals. In 1995 only larvae were analysed.
RAPD-PCR
Reaction buffer was made in 3 mL quantities, sufficient to carry out 60 individual 50 μL reactions. Each reaction consisted of 50 mM KCl, 10 mM Tris-HCl (pH 9.0), 1.5 mM MgCl2, 0.01% gelatin (w/v) and 0.1% Triton X-100, 200 μM dNTPs and 1 μM final primer concentration. Reaction buffer was made up immediately prior to use and unused buffer was discarded. Aliquots (49 μL) were placed in labelled 500 μL microcentrifuge tubes and autoclaved light mineral oil (25 μL) was layered on top. These tubes were placed under UV (260 nm) lamps (Stratagene, LaJolla, CA) for 10 min to destroy contaminating DNA. Mosquito DNA (1 μL) was added through the oil to the mixture. Tubes were placed in the thermal cycler and heated to 95°C for 5 min and the temperature was lowered to 80°C for 20 min during which time 1 unit of Taq DNA polymerase (Promega Biotech) was added to each reaction. Amplifications were carried out at 92°C for 1 min, 35°C for 1 min, slope up to 72°C at 1°C steps every 8 s, 72°C for 2 min. This cycle was repeated 45 times and then held at 72°C for 7 min, the tubes then remained at 4°C until they could be processed.
Twenty-five primers were screened using 8–10 individuals of each species. Duplicate reactions on each individual were used to test the reproducibility of individual bands. Duplicates were amplified on different thermal cyclers run on the same day or in different reaction sets amplified on different days. Primers that did not produce well amplified polymorphic bands that could be clearly distinguished from neighbouring bands were not used. Bands that did not appear in duplicate reactions were not scored. We identified three primers in each species (Table 2) that revealed a large number of well resolved polymorphisms. In all amplifications, a negative control containing all of the reaction components except template DNA was processed.
Gel electrophoresis
PCR products were loaded alongside DNA size markers (1 kb ladder, Gibco, Bethesda Research Laboratories) onto a 1.2% TBE agarose gel containing ethidium bromide. These were subject to electrophoresis for 30 min at 100 V. Bands were visualized on a 302 nm UV light transilluminator to test for amplification. The PCR was repeated on failed amplifications. For the purpose of visualizing and scoring polymorphisms, amplified products were fractionated on 5% polyacrylamide TBE gels and bands were revealed with silver staining (Black & DuTeau, 1997).
Data analysis
We constructed a dataset for each species following the format described in Black (1995). Each line of the dataset contains a code for the collection and an identification number for the individual mosquito followed by a ‘1’ for the presence of a specific band or a ‘0’ for the absence of that band. We scored 47 RAPD-PCR bands in A. cataphylla and A. pullatus and 48 bands in A. hexodontus.
A series of FORTRAN programs were used for population genetic analyses (Black, 1995; Apostol et al., 1996). All programs made three assumptions regarding the inheritance and expression of RAPD polymorphisms among individuals in populations. These were: (i) RAPD bands are inherited as dominant alleles in a Mendelian fashion; individuals with an amplified band are homozygous or heterozygous for a dominant allele whereas individuals without amplified bands are homozygous recessive. (ii) Genotypes are in Hardy–Weinberg proportions; the frequency q of a recessive allele a at a locus is estimated as the square root of the frequency of homozygous recessive individuals and the frequency p of a dominant allele A is 1−q. (iii) Allele A is identical in state among individuals as is allele a. We have studied the inheritance of and mapped the location of RAPD markers in Aedes aegypti (Antolin et al. 1996), verifying the first assumption albeit in another mosquito species. Allozyme genotype frequencies were in Hardy-Weinberg proportions in two studies of breeding structure in other snow pool Aedes (Eldridge et al., 1986; Brust & Munstermann, 1992), suggesting that the second assumption may be valid. The third assumption was not tested.
RAPDFST estimated F-statistics following the methods of Weir & Cockerham (1984) and Lynch & Milligan (1994). RAPDDIST estimated pair-wise Lynch & Milligan (1994) FST values among collections and compiled these into a distance matrix. MANTEL transformed the FST matrix into a matrix of pair-wise FST/(1−FST) values (Rousset, 1997) and transformed (log10) geographical distances (km) among populations and computed the Mantel statistic (Mantel, 1967) between the transformed genetic and geographical distances. MANTEL then performed 1000 random permutations of rows and columns in the geographical distance matrix (Mantel, 1967) and computed the Mantel statistic for each permutation to test if the original correlation is stronger than expected by chance. RAPDDIST also performed a bootstrap analysis by randomly resampling loci from the original dataset to produce a FST matrix for each pseudoreplicated dataset. The distance matrices were then subjected to Unweighted Paired Group Method using Arithmetic Averages (UPGMA) using NEIGHBOR in PHYLIP 3.5C (Felsenstein, 1993) to construct dendrograms and CONSENSE in PHYLIP 3.5C to derive a consensus tree
Results
Figure 2 shows an example of RAPD-PCR polymorphisms detected within and between two snow pools containing Aedes pullatus from Rocky Mountain National Park. Polymorphisms that were scored for genetic analysis are indicated with dots.
RAPD-PCR polymorphisms within and between two Aedes pullatus collections from the Big Thompson River drainage (locations are indicated in Fig. 1). DNA fragments were fractionated in 5% polyacrylamide gels and detected by silver staining. Lanes labelled M are molecular weight markers (Gibco, BRL 1 kb ladder). Dots indicate the loci that were scored, arrows in the margin indicate their size in bp.
FST values were estimated between pairs of pools following Lynch & Milligan (1994). FST/(1−FST) values were plotted against log10 (geographical distances (km)) estimated from GPS coordinates (Table 1). Genetic distances were not correlated with geographical distances in A. hexodontus or A. cataphylla (Fig. 3) and the Mantel test indicated high probabilities that the correlations could have arisen by random chance. Figure 1 indicates some instances where high mountain ranges separate pools that are closer to one another (e.g. Emmaline Lake Trail and Upper Fall River Road (Fig. 1) than pools within the same drainage. Correlations were therefore computed among pools within the same drainage. These correlations were also small and the probabilities from the Mantel test were large (Fig. 3)
Correlation between log10 (geographical distance (km)) and FST/(1−FST) values calculated following Rousset (1997) in three snow pool Aedes species in northern Colorado.
Dendrograms on pair-wise FST values indicate a similar trend (Fig. 4). Collections clustered independently of river drainages or their geographical proximity to one another. For example, the two snow pools containing A. hexodontus in Pingree Park were separated by only 0.1 km but did not cluster together whereas collections from Cirque meadows in the CLP drainage clustered with those from Bierstadt Trail in the BTR drainage.
Cluster analysis of FST values among collections of three snow pool Aedes species in northern Colorado. FST values were calculated following Lynch & Milligan (1994). Trees were constructed from distance matrices with UPGMA analysis with PHYLIP3.5C (Felsenstein, 1993). Numbers above branches indicate the percentage of bootstrap analyses that support that branch. Percentages are only provided on those branches with >50% support.
After observing this trend in 1994, we decided to examine the same pools in 1995 to examine gene flow among local pools. There were seven pools in which we were able to make large collections of the same species in 1995. In A. pullatus the 1994 and 1995 collections from the Upper Fall River Road (elev. 3523 m) clustered together with 89% bootstrap support. Similarly 1994 and 1995 collections of the same species from the Emmaline Lake Trail (elev. 3244 m) clustered together with 76% bootstrap support. Although 1994 and 1995 collections of A. pullatus from the Signal Peak Trailhead (elev. 2602 m) clustered together, bootstrap support was less than 50%. Two of the three collections of A. hexodontus made in consecutive years clustered together but none clustered with high bootstrap support. We had only one pool where A. cataphylla could be collected in both years (Fern Lake Trail, elev. 2486 m); these pools clustered with 98% bootstrap support.
Estimation of Nm
The observed patterns of breeding structure clearly violate two key assumptions of Wright's Island Model used to estimate effective migration rate Nm from FST with the equation:

Genetic drift appears to account for most of the genetic differentiation among collections within a drainage and bootstrap analysis of the same pool across years suggests that Nm is not uniform among all snow pools in the study. Nevertheless, we estimated Nm from FST as a means to compare effective migration rates among species.
Estimates of θ (Weir & Cockerham, 1984) were 0.059 for A. cataphylla, 0.033–0.061 for A. hexodontus and 0.058–0.099 for A. pullatus and jackknife estimates of the standard error around θ were generally small (Table 3). FST values estimated by the method of Lynch & Milligan (1994) were similar. Average Nm values were 3.5–4.0 for A. cataphylla, 5.8–5.9 for A. hexodontus and 3.2–3.4 for A. pullatus. These results suggest that A. cataphylla and A. pullatus have slightly lower Nm than A. hexodontus. FST values in Fig. 4 are plotted at the same scale. Note that FST among A. hexodontus and A. cataphylla snow pools is less than those among pools of A. pullatus.
Discussion
FST values between pools within drainages were independent of geographical distance in A. cataphylla and A. hexodontus and, in all three species, pools clustered across drainages and generally independently of geographical proximity. These observations suggest two patterns. First, larvae in proximate snow pools become randomly genetically differentiated through drift. Secondly, within the 50 km range examined within this study, mountains and broad valleys in northern Colorado do not in the long term act as major barriers to gene flow among snow pool Aedes. However, these species are distributed throughout the United States and Canadian Rocky Mountains and occur north through the southern half of Alaska. In addition, A. hexodontus is distributed across northern Canada and A. pullatus has a disjunct population in northern Quebec province (Darsie & Ward, 1981). At these greater geographical scales, isolation by distance is very likely in A. hexodontus and A. cataphylla and should increase over that observed in this study of A. pullatus.
These patterns are consistent with an hypothesis in which females in spring mate, feed and oviposit at or near their emergence sites but later in the season are forced to disperse to higher altitudes in search of blood meals and oviposition sites. Limited Nm among adults in spring could be one of the causes of the genetic drift observed among local pools and explain the clustering of pools in consecutive years. Larvae in pools would remain isolated and drift apart if there is limited Nm among adults from the site of larval development.
Low temperatures decrease the rate of snow melt and increase larval development time in high altitude pools of A. pullatus and might limit Nm among adults emerging from these sites. This could cause the clustering of 1994 and 1995 collections of A. pullatus at Upper Fall River Road and Emmaline Lake Trail. Adults from these pools hatch in early August and have from 6 to 8 weeks to mate, blood feed and lay eggs before frost and snow return in mid-late September. In contrast, adult A. pullatus first appear in early June at the Signal Peak Trailhead pool (2600 m). 1994 and 1995 collections at this pool also clustered together but without strong bootstrap support.
Significant clustering was also observed with 1994 and 1995 collections of A. cataphylla. Larvae of this species appear as soon as snow melts in early March; they survive repeated freezing and adults are often in flight by early May (Harmston & Lawson, 1967). It may be that the severe weather conditions in early spring limit Nm among adults such that females mate, feed and lay eggs locally. Most A. cataphylla larval habitats are dry by the end of May. Seasonal phenology may therefore serve to limit effective migration among A. cataphylla adults.
Aedes hexodontus larvae are distributed ubiquitously along the banks of rivers in both BTR and CLP drainages, which should lead to greater gene flow. In contrast, A. pullatus and A. cataphylla larval habitats tend to be spatially isolated in our study locations. FST values were larger and Nm smaller in A. pullatus and A. cataphylla.
The more continuous distribution of A. hexodontus might lead to isolation by distance among pools in this species. However, there are a number of factors that could contribute to the apparent random association observed among A. hexodontus collections and the lack of significant clustering among collections in consecutive years. Aedes hexodontus larval habitats occur along rivers where whole pools are washed away during spring run-off. For example, pools containing early larval instars were observed in early April 1995 at Alluvial Fan Park and Pingree Park II pools. But these were washed away as the stream left its bank during run-off. In addition, we observe high larval mortality in A. hexodontus larvae resulting from microsporidian infections (Andreadis, 1983; Sweeney & Becnel, 1991). This mortality becomes extremely high in late May and early June as larval densities increase in drying pools. Random mortality would explain why two of the three 1994–95 collections of A. hexodontus clustered together but neither had strong bootstrap support.
Adult females of all three species survive well into late summer. Carpenter & Nielsen (1965) reported collecting biting A. cataphylla in mid July and A. hexodontus in late August in northern Utah. We have collected adults of all three species at or above 4000 m through late August. Mosquitoes active in late summer are likely to face a paucity of oviposition sites and sources of blood. Although oviposition behaviour in snow pool Aedes is not well known, humidity seems to be the determining factor in site selection (Iversen, 1971). Most larval habitats are temporary pools that are dry by mid-summer. Female mosquitoes probably find oviposition sites harder to recognize after the sites become dry. Furthermore, snow pools attract vertebrate hosts that serve as sources of blood meals. Hosts will also therefore become less abundant as pools dry in mid-late summer.
The maximum flight range of these mosquitoes has not been well explored in the mountains of Colorado, but in other regions of the western US (Nielsen, 1957) and in the subarctic (Blanton et al., 1950) they appear to be capable of flying great distances. As their emergence sites disappear, females may seek remaining pools that retain water late in the summer. This may result in dispersal over great distances and at higher altitudes. As the summer progresses only the rare pools that still contain water will receive migrants seeking oviposition sites. These more permanent pools may serve as reservoirs for migrant eggs in future generations.
References
Andreadis, T. G. (1983). An epizootic Amblyospora sp. in field populations of the mosquito, Aedes cantator. J Invert Pathol., 42: 427–430.
Antolin, M. F., Bosio, C. F., Cotton, J., Sweeney, W. P. and Black, W. C., I. V. (1996). Rapid and dense linkage mapping in a wasp (Bracon hebetor) and a mosquito (Aedes aegypti) with Single Strand Conformation Polymorphisms Analysis of Random Amplified Polymorphic DNA markers. Genetics, 143: 1727–1738.
Apostol, B. L., Black, W. C., I. V., Reiter, P. and Miller, B. R. (1996). Population genetics with RAPD-PCR markers: the breeding structure of Aedes aegypti in Puerto Rico. Heredity, 76: 325–334.
Baker, M. (1961). The altitudinal distribution of mosquito larvae in the Colorado Front Range. Trans Am Ent Soc, 87: 231–246.
Black, W. C., I. V. (1995). Statistical analysis of arbitrarily primed PCR patterns in molecular taxonomic studies. In: Clapp, C.L. (ed.) Methods in Molecular Biology, vol. 50, Species Diagnostics Protocols: PCR and Other Nucleic Acid Methods, pp. 39–55. Humana Press, Totowa, NJ.
Black, W. C., I. V. and Duteau, N. M. (1997). RAPD-PCR and SSCP analysis for insect population genetic studies. In: Crampton, J., Beard, C. B. & Louis, C. (eds) The Molecular Biology of Insect Disease Vectors: A Methods Manual, pp. 361–373. Chapman and Hall, New York.
Blanton, F. S., Travis, B. V., Smith, N. and Husman, C. N. (1950). Control. of adult mosquitoes in Alaska with aerial sprays. J Econ Entomol, 43: 347–350.
Brust, R. A. and Munstermann, L. E. (1992). Morphological and genetic characterization of the Aedes communis complex (Diptera: Culicidae) in North America. Ann Entomol Soc Am, 85: 1–10.
Carpenter, M. J. and Nielsen, L. T. (1965). Ovarian cycles and longevity in some univoltine Aedes species in the Rocky Mountains of western United States. Mosquito News, 25: 127–134.
Darsie, R. F., J. R. and Ward, R. A. (1981). Identification and geographical distribution of the mosquitoes of North America, north of Mexico. Mosquito Syst (Suppl), 1: 1–313.
Edwards, F. W. (1932). Diptera, Family Culicidae. In: Wytsman, P. (ed.) Genera Insectorum. Fasc. 194. Tervuren, Belgium.
Eldridge, B. F., Munstermann, L. E. and Craig, G. B. (1986). Enzyme variation in some mosquito species related to Aedes (Ochlerotatus) stimulans (Diptera: Culicidae). J Med Entomol, 23: 423–428.
Ellis, R. A. and Brust, R. A. (1973). Sibling species delimitation in the Aedes communis (DeGeer) aggregate (Diptera: Culicidae). Can J Zool, 51: 915–959.
Felsenstein, J. (1993). PHYLIP: Phylogeny inference package, version 3.5C. University of Washington, Seattle, WA.
Hardy, J. L., Eldridge, B. F., Reeves, W. C., Schutz, S. J. and Andpresser, S. B. (1993). Isolations of Jamestown Canyon virus (Bunyaviridae: California Serogroup) from mosquitoes (Diptera: Culicidae) in the western United States, 1990–92. J Med Entomol, 30: 1053–1059.
Harmston, F. C. and Lawson, F. A. (1967). Mosquitoes of Colorado. U.S. Department of Health, Education and Welfare/ Public Health Service, Atlanta, GA.
Iversen, T. B. (1971). The ecology of a mosquito population (Aedes communis) in a temporary pool in a Danish beech wood. Arch Hydrobiol, 69: 309–332.
Livak, K. (1984). Organization and mapping of a sequence on the Drosophila melanogaster X and Y chromosomes that is transcribed during spermatogenesis. Genetics, 107: 611–634.
Lynch, M. and Milligan, B. G. (1994). Analysis of population genetic structure with RAPD markers. Mol Ecol, 3: 91–99.
Mantel, N. (1967). The detection of disease clustering and a generalized regression approach. Cancer Res, 27: 209–220.
Nielsen, L. T. (1957). Notes on the flight ranges of Rocky Mountain mosquitoes of the genus Aedes. Proc Utah Acad Arts Sci Letts, 34: 27–29.
Ross, H. H. (1964). The colonization of temperate North America by mosquitoes and man. Mosquito News, 24: 103–118.
Rousset, F. (1997). Genetic differentiation and estimate of gene flow from F-statistics under isolation by distance. Genetics, 145: 1219–1228.
Sweeney, A. W. and Becnel, J. J. (1991). Potential of microsporidia for the biological control. of mosquitoes. Parasit Today, 7: 217–220.
Weir, B. S. and Cockerham, C. C. (1984). Estimating F-statistics for the analysis of population structure. Evolution, 38: 1358–1370.
Williams, J. G. K., Kubelik, A. R., Livak, K. J., Rafalski, J. A. and Tingey, S. V. (1991). DNA polymorphisms amplified by arbitrary primers are useful genetic markers. Nucl Acids Res, 18: 6531–6535.
Wood, D. M., Dang, P. T. and Ellis, R. A. (1979). The Insects and Arachnids of Canada, Part 6. The Mosquitoes of Canada. Agric. Canada Publication no. 1686, Ottawa, Canada.
Acknowledgements
We thank the National Park Service for permission to collect in Rocky Mountain National Park and Debesh West, Chris Bosio, and Doug Norris for collecting larvae. This research was supported by the MacArthur Foundation Network on the Biology of Parasite Vectors and by a National Institutes of Health Grant AI34454.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
West, D., Black IV, W. Breeding structure of three snow pool Aedes mosquito species in northern Colorado. Heredity 81, 371–380 (1998). https://doi.org/10.1046/j.1365-2540.1998.00387.x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1046/j.1365-2540.1998.00387.x