Abstract
The chromosomal radiation of the house mouse in the island of Madeira most likely involved a human-mediated colonization event followed by within-island geographical isolation and recurrent episodes of genetic drift. The genetic signature of such processes was assessed by an allozyme analysis of the chromosomal races from Madeira. No trace of a decrease in diversity was observed suggesting the possibility of large founder or bottleneck sizes, multiple introductions and/or a high post-colonization expansion rate. The Madeira populations were more closely related to those of Portugal than to other continental regions, in agreement with the documented human colonization of the island. Such a Portuguese origin contrasts with a study indicating a north European source of the mitochondrial haplotypes present in the Madeira mice. This apparent discrepancy may be resolved if not one but two colonization events took place, an initial north European introduction followed by a later one from Portugal. Asymmetrical reproduction between these mice would have resulted in a maternal north European signature with a nuclear Portuguese genome. The extensive chromosomal divergence of the races in Madeira is expected to contribute to their genic divergence. However, there was no significant correlation between chromosomal and allozyme distances. This low apparent chromosomal impact on genic differentiation may be related to the short time since the onset of karyotypic divergence, as the strength of the chromosomal barrier will become significant only at later stages.
Similar content being viewed by others
Introduction
Oceanic islands have long attracted the attention of biologists in their endeavour to document the evolutionary processes involved in adaptive radiations. A renewed interest has recently focused on Atlantic archipelagos where several discrete groups of small islands may harbour a large diversity of habitats. Such a situation provides an ideal framework for unravelling the interaction between landscape features and microevolutionary processes leading to species diversification (Emerson and Kolm, 2005; Gavrilets and Vose, 2005). However, ascertaining the origin and evolution of island species requires that the number of colonization events be determined, the time of the radiation estimated and the continental source of founder populations identified. Recent improvements in phylogenetic reconstruction and meta-analyses have provided powerful tools to tease apart the factors promoting adaptive radiations and endemism (Emerson, 2002; Manel et al., 2003). The proportion of endemic species in particular has been shown to be strongly related to island size and topographical complexity due to the increased opportunity for intra-island allopatric speciation (Emerson and Kolm, 2005). Recently, such approaches have been successfully applied to various plant and animal species diversifications in the Macaronesian islands (Jordal and Hewitt, 2004; Salgueiro et al., 2004; Silvertown, 2004).
More recent within-species radiations can also be detected. A case in point is the impressive chromosomal radiation discovered in the house mouse in Madeira (Britton-Davidian et al., 2000). This island is part of an Atlantic volcanic archipelago consisting of Madeira, Porto Santo and Desertas, all of which are Portuguese dependencies. They are relatively small and flat islands with the exception of Madeira in which sharp changes in altitude are present. The karyotypic analysis of mice from the Madeiran archipelago uncovered the presence of six highly differentiated chromosomal races carrying different combinations of Robertsonian (Rb) centric fusions between acrocentric chromosomes (Britton-Davidian et al., 2000). Although such rearrangements are widespread in mouse populations from continental Europe and North Africa (Piálek et al., 2005), 10 of the 20 Rb fusions discovered are unique to this archipelago. The chromosomal differentiation was observed in Madeira, whereas the other islands were found to have mice carrying the standard 2n=40 all acrocentric karyotype (Mathias et al., 2004). A previous study postulated that this radiation was related to the extreme topography of Madeira, which isolated mouse populations, providing the necessary conditions for the recurrent fixation of Rb fusions through genetic drift (Britton-Davidian et al., 2000). Not only is the extent of the chromosomal radiation in Madeira house mice extraordinary, but also its rate. Historical documents date the first human occupation of Madeira at the end of the fifteenth century AD by Portuguese settlers, suggesting that mice could have been introduced at that time (Britton-Davidian et al., 2000). Alternatively, it has been suggested that Madeira was colonized in the ninth century AD by Danish Vikings; this suggestion follows from a similarity in mouse mitochondrial DNA sequence between individuals in northern Europe and the Madeiran archipelago (Gündüz et al., 2001). Whatever the origin, such dates provide very high rates of Rb fusion fixation (9 in 500–1000 years; Britton-Davidian et al., 2005).
The stochastic factors that apparently promoted chromosomal race formation in Madeiran mice might be expected to leave a genetic signature (Frankham, 1997), such as a within-race loss of diversity and a high between-race genic divergence. This study presents an allozyme analysis of house mouse populations from Madeira and the second largest island in the archipelago, Porto Santo to (1) trace the effects of genetic drift by estimating genic diversity within populations, (2) determine the effects of geographic isolation and chromosomal differentiation on the genic divergence between populations and races and (3) identify the origin of the founder populations by comparing the island populations to samples from Portugal and Denmark as well as a large published geographical survey of other European and North African house mice. These analyses provide a preliminary assessment of the evolutionary processes that may be involved in the chromosomal radiation of house mice in Madeira.
Materials and methods
Population samples
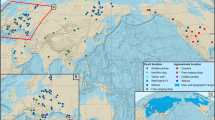
Mice belonging to Mus musculus domesticus were captured in commensal habitats (houses, gardens, farms and cultivated fields) using Sherman live traps baited with sardine paste. One hundred and fifty-eight mice from 37 localities in Madeira (1998 and 1999 field trips) were analyzed together with 24 mice from seven localities in Porto Santo. The specimens from Madeira derived from the different chromosome races (see Table 1) and the chromosomal hybrid zone between the Santana race and standard mice in Funchal, the main port of entry onto the island (see Britton-Davidian et al., 2000, 2005). In addition, nine mice were trapped in two localities from central Portugal (Caixas 38°40N 9°20W and Santarém 39°23N 8°68W). Most of these localities were pooled according to geographical and/or chromosomal proximity into 22 population samples (17 in Madeira, 4 in Porto Santo and 1 in continental Portugal; see Figure 1). For comparative purposes, specimens of M. m. domesticus were trapped along the eastern coast of southern Denmark in May 2000 (see details in Smadja et al., 2004). Several farmhouses were sampled around two main towns yielding 17 mice in Sommersted (Kastvra A, Ringtved, Sillerupgaard, Kastvra) and 21 mice in Sonder Bjert (Skartved, Binderup, Soender Stenderup, Lauritzminde). The names of the island Rb races follow the nomenclature proposed by Piálek et al. (2005) except that the prefix P (for Portugal) was omitted for brevity.
Maps of Madeira and Porto Santo. Numbers refer to the sampled localities and the dashed line indicates those pooled per population and per race. San=Santana, Asj=Arco di San Jorge, Pod=Ponta Delgada, Svi=Sao Vicente, Edc=Estreito de Calheta, Adc=Achadas da Cruz, Adc/Edc=hybrid population between the Adc and Edc races, Ldb=Lugar di Baixo, Psa=Porto Santo, Fun=hybrid zone of Funchal. The insert shows the distribution of localities in Funchal. Distribution of Rb fusions among the races: San (including Asj)=2.19, 3.8, 4.16, 5.14, 6.7, 9.10, 11.12, 13.17, 15.18; Pod=3.8, 4.5, 9.18, 10.16, 11.12, 14.17; Svi=2.4, 3.8, 5.18, 6.7, 9.12, 10.16, 11.19, 13.17; Edc=2.4, 3.14, 5.18, 6.7, 8.11, 9.12, 10.16, 13.17; Adc=2.4, 3.14, 5.18, 7.15, 8.11, 9.12, 10.16, 13.17; Ldb=2.4, 3.14, 6.7, 8.11, 9.12, 10.16, 13.17, 15.18.
The karyotypes of the mice from Madeira, Porto Santo, mainland Portugal and Denmark have all been previously described (Gazave et al., 2003; Mathias et al., 2004; Smadja et al., 2004; Britton-Davidian et al., 2000, 2005; Nunes et al., 2005).
Allozyme analysis
The allozyme analysis was performed on 229 mice. A total of 33 loci were scored from liver, kidney, heart, plasma and hemolysate samples by starch gel electrophoresis (see Britton-Davidian, 1990 for references on methods): alcohol dehydrogenase ADH (1.1.1.1, Adh-1), albumin ALB (Alb-1), amylase AMY (3.2.1.1, Amy-1), carbonic anhydrase CAR (4.2.1.1, Car-2), creatine kinase CK (2.7.3.2, Ck-1), esterases ES (3.1.1.1, Es-1, Es-2, Es-3, Es-10, Es-15), guanine deaminase GDA (3.5.4.3, Gda-1), α-glycerophosphate dehydrogenase αGPD (1.1.1.8, Gdc-1), glyoxalase GLO (4.4.1.5, Glo-1), amino-aspartate transaminases GOT (2.6.1.1, Got-1, Got-2), glycerophosphate isomerase GPI (5.3.1.9, Gpi-1), hydroxyacid oxidase HAO (1.1.3.15, Hao-1), haemoglobin HBB (Hbb), isocitrate dehydrogenase IDH (1.1.1.42, Idh-1, Idh-2), lactate dehydrogenase LDH (1.1.1.27, Ldh-1, Ldh-2), NADP-dependent malate dehydrogenases MOD (1.1.1.40, Mod-1, Mod-2), NAD-dependent malate dehydrogenase MOR (1.1.1.37, Mor-1, Mor-2), mannose phosphate isomerase MPI (5.3.1.8, Mpi-1), nucleoside phosphorylase NP (2.4.2.1, Np-1), phosphoglucomutase PGM (5.4.2.2, Pgm-1, Pgm-2), pyruvate kinase PK (2.7.1.40, Pk-1, Pk-3) and superoxide dismutase SOD (1.15.1.1, Sod-1). The identity and nomenclature of alleles were assessed by comparison with the migration profiles of the alleles present in two laboratory strains of mice (C57Bl/6J and Balb/c; see Britton-Davidian et al. (1989) for details of the procedure).
Statistical analyses
Allele frequencies were estimated as well as the following measures of intrapopulation variation: mean number of alleles (A), percentage of polymorphic loci per sample (P%) and mean expected heterozygosity (He) calculated for small samples using the BIOSYS-1 program (Swofford and Selander, 1981). All other population parameters were computed and tested as implemented in GENEPOP v3.2a (Raymond and Rousset, 1995) unless otherwise indicated. In all cases, the default options were used. Exact tests with their associated P-values were performed in each population to test for departure from Hardy–Weinberg expectations and for pairwise linkage disequilibrium between loci. Genic differentiation between pairs of populations was estimated by Fisher's exact test. In addition, pairwise FST values using the estimator θ were calculated, and their level of significance evaluated by permutation tests (1000) using GENETIX v4.03 (Belkhir et al., 1996–2004). Relationships between populations were investigated by constructing a maximum likelihood (ML) tree from the gene frequency data, and a distance tree using the neighbour-joining method (NJ). The genetic distance used was the FST-based Reynolds' index available in GENETIX v4.03. Both trees were built with the PHYLIP v3.5c package (Felsenstein, 1985). Branch support was established by 500 bootstrap resamplings among loci. The graphical display of the tree topologies was drawn with the TreeView v1.6.6 software distributed by Page (1996). The same procedures were applied to the chromosomal data by considering each Rb fusion as a locus, with the acrocentric and metacentric states treated as alternative alleles. In this case, two additional tests were performed: linkage disequilibrium between Rb fusions and allozyme loci and a test of the correlation between genetic distances (Nei, 1978) for Rb fusions vs those for allozymes using the Mantel procedure as implemented in GENETIX v4.03 (1000 permutations). P-values for the multiple tests were subjected to sequential Bonferroni correction.
To investigate the genetic relationship of the island populations to Portugal and Denmark, a Bayesian analysis of population structure was performed as implemented in BAPSv.4.14 (option: Trained Mixture Analysis using these two European samples as the reference file; Corander et al., 2006). To compare genic diversity and relatedness of the populations from the Madeira archipelago over a larger European sample, allozyme data from an additional 27 European and North African house mouse populations (Britton-Davidian et al., 1989; Britton-Davidian, 1990) were included in the analyses. He estimates, genetic distances and phenetic trees were assessed over the 31 loci common to all samples (Haox and Gda were omitted). Differences in He between geographical groupings of populations were tested by the non-parametric Kruskal–Wallis test. Population genetic structure was investigated by analyses of molecular variance (AMOVA, SAMOVA), which estimate the proportion of genic variation at different hierarchical levels using information from the geographical distribution of allozyme frequencies and the pairwise distance between them. Two approaches were performed. The first one involved an AMOVA (Excoffier et al., 1992) of user-established hierarchical geographical arrangements of populations following the clusters present in the NJ tree. The procedure consisted of a step-by-step increase in the size of the geographical groups to uncover those with the maximum among-group divergence. The second series of tests was implemented with the variant spatial analysis of molecular variance (SAMOVA) approach (Dupanloup et al., 2002) which detects the groups of populations that are maximally differentiated from each other. As in the AMOVA procedure, the proportion of total genic variation due to differences between groups of populations is maximized within a sequentially decreasing number of groups.
Results
Chromosomal diversity
Details of the chromosomal composition of mice from the islands of Madeira and Porto Santo are published in Britton-Davidian et al. (2005) and Nunes et al. (2005), of the Portuguese sample in Mathias et al. (2004) and of the Danish populations in Smadja et al. (2004). In summary, six chromosomal races harbouring a different combination of 20 Rb fusions are present on Madeira (Figure 1; Asj is considered as belonging to the Santana Rb race, see below). No hybrid karyotypes between Rb races were observed except in one locality where two mice were trapped each carrying Rb fusions that discriminate the westernmost races (Achadas da Cruz race/Estreito da Calheta race (Adc/Edc)). Within-race polymorphism involving Rb fusions and their homologous acrocentrics was found in four of the six races as well as in the hybrid zone localities in Funchal. The mice from mainland Portugal and Porto Santo had the 2n=40 standard karyotype with the exception of two individuals in the latter island, each heterozygous for a different Rb fusion also present in Madeira. The house mice from Denmark showed a diploid number varying from 2n=34 to 37 due to the presence of three Rb fusions: Rb(3.8), Rb(2.5) and Rb(6.9). The latter two were segregating in Den1 whereas only Rb(2.5) was polymorphic in Den2. The levels of chromosomal heterozygosity (Hk) in these populations are reported in Table 1. All Hk values were low, except for most of the samples in the hybrid zone of Funchal (maximum of 0.21 in Fun4) and Den1 (0.25). Heterozygosity for multiple Rb fusions (2–8) followed the same trend, as it was almost exclusively limited to samples from Funchal, particularly Fun4 in which one individual was heterozygous for 8 Rb fusions. No significant departures from Hardy–Weinberg expectations or linkage disequilibrium between Rb fusions were observed.
Genic diversity
The allozyme analysis of house mice from Madeira, Porto Santo, Portugal and Denmark showed that eight of the 33 loci scored were monomorphic for the same allele in all populations (Appendix A). The remaining 25 loci were polymorphic in at least one population. Significant deviations from Hardy–Weinberg expectations were detected in only three localities: Fun2 (1 locus; P<0.01), Fun4 (2 loci; P<0.05) and Adc1 (1 locus; P<0.01). In all three cases, deviations were due to a deficit of heterozygotes involving three loci (Car, Es-15, Gda). Linkage disequilibrium between pairs of loci was tested within all populations. Differences were found between one to seven pairs of loci in seven of the 22 populations. However, given the large number of tests involved, we applied the Bonferonni correction and none remained statistically significant.
Measures of genic diversity are provided in Table 1. Estimates of expected heterozygosity varied between localities; however, the highest values were present in Madeira where they ranged from 0.16 in the hybrid zone (Funchal) to 0.08 in Pod. The lowest values were recorded in the Danish samples (mean He=0.05). These estimates were compared to previously published values for 27 populations from Europe and North Africa (Britton-Davidian et al., 1989; Britton-Davidian, 1990), recalculated over the 31 loci common to both studies. To account for variation between northern and southern European populations and between Madeira and Porto Santo, He values were subdivided into four geographic groups. Mean values between these regions showed that He was highest in the Rb races from Madeira (even when Funchal was excluded) and southern European populations compared to Porto Santo and northern European populations (Table 2). These differences were marginally significant (Kruskal–Wallis test; P=0.048). Post hoc tests between regions indicated that the differences were significant only between the Madeiran races and the north European populations. Twelve unique alleles were observed in the populations studied (see Appendix A). None were present in Porto Santo and only three in the Madeiran Rb races, two in Svi and one in Ldb. Elsewhere unique alleles were evenly distributed between the Portuguese (3), Danish (3) and Funchal (3) samples.
Genic differentiation within the Madeiran archipelago
Population differentiation was investigated within and between races and between islands in the Madeiran archipelago. The exact tests of genic differentiation involving the Rb races detected no significant divergence between samples within races except for the Santana race (San1 vs San2: Table 3). Also the Achadas da Cruz race (Adc) and the Estreito da Calheta race (Edc) samples were not significantly differentiated from the hybrid interracial sample (Adc/Edc; P>0.16). Piálek et al. (2005) recognized Asj (Arco di San Jorge) as a seventh race distinct from the Santana race, since it lacked one of the Rb fusions present in the latter. However, as several mice carrying this fusion were subsequently trapped in the Asj localities, this distinction is no longer warranted (Britton-Davidian et al., 2005). The relatedness between mice from Asj and San is now further supported by the absence of a significant genic differentiation between these samples (P>0.60). In Porto Santo, the four localities sampled were slightly differentiated (P<0.05). Pairwise tests within the hybrid zone in Funchal yielded varying results with Fun1 showing the lowest level of differentiation (P>0.19) from all the other samples except Fun6, which was highly divergent (P<0.001). The level of differentiation between races in Madeira was recalculated after pooling samples within races, as well as in Porto Santo and Denmark (Table 3). Results indicated that most Rb Madeiran races were genically differentiated from each other, with the exception of San vs Svi and Ldb, Ldb vs Svi and Adc vs Edc. Levels of differentiation were then tested between the Madeiran Rb races, Funchal and the Porto Santo mice, as well as between these and the mainland populations (Portugal and Denmark) and were found to be highly significant in all cases (0.001<P<0.01).
To investigate the contribution of chromosomal divergence to genic differentiation, a correlation between genic and chromosomal distance using Nei's genetic distances was performed for the Madeira Rb races and found to be marginally non-significant (Mantel test z=0.75±0.01, P=0.08). In fact, the most chromosomally divergent races (San and Adc) were not among the most genically differentiated (P<0.01).
Island–mainland genic divergence
Interpopulation divergence estimated by the FST index showed three levels of differentiation (Table 3). First, FST values were the lowest among the Madeira Rb races ranging from 0.025 to 0.363 (P>0.05 in 6 cases out of 15), with Pod showing the highest overall levels (0.146–0.363; all P<0.05). Second, the Madeira Rb races were less differentiated from the Portuguese population (0.087–0.292; 0.05>P>0.05), than they were from the Porto Santo (0.279–0.476; all P<0.05) or Danish populations (0.187–0.392; all P<0.05). Third, the divergence between the latter two yielded the highest value observed (0.487; P<0.05). The hybrid samples of Funchal diverged less from the various Madeiran Rb races and the Portugal population, than from the mice of Porto Santo and Denmark. To further evaluate the significance of these genic affinities, an analysis of population structure using a Bayesian approach (BAPS) was performed over all samples except the hybrid population from Funchal. The analysis provided an optimal partition of the data into three clusters using the Portugal and Denmark samples as the reference populations (log (marginal likelihood)=−1265.3143). These groups are shown in Table 4, which includes the log(ml) values associated with changes in cluster assignment of samples. The island populations were distributed into the following three clusters: (1) one including all the Madeira races except Pod (Mad5; Table 4), which formed a separate group, although the low log(ml) value of −1.1 indicated that Pod could also be reasonably allocated to the Madeira Rb group (see BAPSv4.14) and (2) one showing the divergent position of the Porto Santo populations which were always separate from the Madeira ones. In addition, the data showed that a reassignment of the Rb populations to the Portuguese group yielded a less worse partition than when included in the Denmark cluster, except again for Pod which had low and similar values in each case.
The distinctiveness of the Porto Santo mice prompted us to determine the origin and genetic relatedness of the populations studied over a larger geographical sample of house mice. Reynold's distances between these and an additional 27 populations were computed and used to construct an ML tree (Figure 2). The topology highlighted several features. First, the Madeira Rb races clustered together as well as with Portugal. All of these in turn were included in a larger group with the populations from Israel, Egypt and Denmark and one from Tunisia (Tu1). Second, the mice from Porto Santo clustered with the samples from Scotland (Gb1) and France (Fr6) in a composite group including mostly western Mediterranean samples. Third, house mice showed a very low overall macrogeographical genetic structure except at a regional local scale. This weak structure is confirmed by the very low bootstrap values (only three were above 50%) providing little support for the different nodes. The NJ tree topology (not shown) roughly matched what was observed with the ML tree, the only notable difference concerning the position of the Madeiran Pod race, which clustered with the populations from Denmark.
Maximum likelihood tree showing the phylogenetic relatedness of the island and European samples based on the 31 loci common to this and previous studies (Britton-Davidian et al., 1989; Britton-Davidian, 1990). Boostrap values above 50% are indicated in italics. Circled numbers refer to the groups tested in the AMOVA analysis (see text). Key to populations (including sample sizes in the case of the 27 new populations): Algeria (Al1=Bouzadjar 12, Al2=Oran 8), Egypt (Eg1=GZA 18, Eg2=CRO 20), France (Fr1=ENSAM 7, Fr2=Pratx 24, Fr3=Pagès 6, Fr4=Haute Savoie 9, Fr5=Toulouse 10, Fr6=Dordogne 24), Germany (Ge1=Ravensburg 10, Ge2=Tubingen2 12, Ge3=Tubingen3 9), Great Britain (Gb1=CTC 26), Greece (Gr1=Prosotsani 27, Gr2=Orestias 9, Gr3=Marasia 15, Gr4=Doirani 15), Israel (Is1=Geshum 5, Is2=Elrom 10), Italy (It1=Santa Caterina 27, It2=Ovada 22, It3=Bergamo 30), the Netherlands (Ne1=Amsterdam 9), Spain (Sp1=RPT, AVY, VDX 10, Sp2=LRA, MOY 13), Tunisia (Tu1=Sfax 10). Bold: Denmark (Den1=Sommersted, Den2=Sonder Bjert), Madeira (San=Santana, Pod=Ponta Delgada, Svi=Sao Vicente, Edc=Estreito de Calheta, Adc=Achadas da Cruz, Ldb=Lugar di Baixo), Porto Santo (Psa1 Psa2, Psa3/4) and Portugal (Por=Caixas). For further details of the European and Mediterranean samples, see Britton-Davidian et al. (1989) and Britton-Davidian (1990). log-likelihood=2754.07.
To test the significance of the genetic units uncovered by the tree, the partitioning of variation between and within the five groups was assessed by the AMOVA test (see node indication in Figure 2). Results of the analysis did not provide statistical support for these units since the among-group differentiation was lower than that within groups (FCT=0.205 and FSC=0.267, respectively). The SAMOVA method that maximizes FCT by constraining the number of groups but not their composition was then performed. The program was instructed to identify from 10 to 3 partitions of the data (see Table 5 for group compositions). Several geographic clusters were stable throughout the analyses: Portugal with the Madeiran samples, Scotland with Dordogne (Fr6) and one to all populations from Porto Santo, a western Mediterranean group (Italy, Spain with all but one of the French populations), an eastern Mediterranean group (Israel with Egypt) and finally a German group. Thus, this analysis provided statistical support for several of the branching arrangements uncovered in the ML tree. In all analyses using less than seven partitions, among-group differentiation fell below that of within groups. Additional tests were run by removing the North African (Al2, Tu1) and Ne1 (Amsterdam) outlier samples, the group assignment of which fluctuated considerably between tests. The lowest number of significantly differentiated groups decreased to four yielding approximately the same genetic clusters as in the previous seven-group test except for the Danish populations which were now included together with the eastern Mediterranean ones in the Madeira/Portugal group (FSC=0.236 and FCT=0.249; Table 5). In no instance did the Madeira Rb races cluster with the Danish samples alone, nor with any other European/Mediterranean population. The analyses of genetic variance thus provided added support for a different geographic make-up of the mice from Porto Santo and Madeira, as well as the Portuguese affinity of the latter.
Discussion
Genic diversity and colonization history
Population genetic models predict that accidental colonization of islands often involves a small number of individuals leading to a loss of genic variation and an instantaneous increase in genic divergence from ancestral populations (Frankham, 1997; Berry and Scriven, 2005). Two other specific features may be expected to enhance these effects in Madeira house mice: the small size of the island with its highly dissected topography, and the subdivision of the mouse population into chromosome races. However, the results we obtained were quite the contrary. The allozyme analysis shows that the Rb races exhibit no decrease in genic diversity compared to European populations; indeed they are in fact among the highest levels recorded in the house mouse (Britton-Davidian et al., 1989; Britton-Davidian, 1990). A further indication lies in the lack of a reduction in the mean number of alleles which is more sensitive than He to bottlenecks (Frankham, 1997). On the other hand, very few unique alleles are present in these races. Theoretical models predict that recovery in genic heterozygosity following a bottleneck is caused by new mutations which slowly increase in frequency by genetic drift (Nei, 2005). If unique alleles are indicative of the occurrence of novel mutations, their low number in the Madeiran Rb races compared to the mainland populations may reflect the short time since the colonization event which can be roughly estimated at 2000 generations for a colonization from Portugal (500 years and four generations/year; see Nachman and Searle, 1995) and 4000 generations for the earlier suggested colonization from northern Europe. Conversely, a slight reduction in all diversity parameters is found in the mice of Porto Santo in which no chromosomal differentiation has taken place. A reduced level of variability is also present in the mice from Denmark, and is in agreement with a gradual loss of diversity due to founder events during the South to North expansion of the house mouse in Europe (Britton-Davidian, 1990; Nachman et al., 1994).
Several non-mutually exclusive reasons may explain the pattern found in the Madeira mice: (1) a large and geographically varied introduction source, (2) a high post-colonization expansion rate (Zenger et al., 2003) and (3) sufficiently large founding populations to prevent post-introduction losses in diversity. Previous data from house mouse populations indicate that diversity levels in island populations are sometimes higher than in the mainland (Navajas y Navarro and Britton-Davidian, 1989). This finding can be attributed to the extraordinary level of passive dispersal of house mice as a commensal of humans. Such a trait will facilitate multiple introductions onto islands from different sources, thereby increasing local variation (for example, Mediterranean island populations; Navajas y Navarro and Britton-Davidian, 1989). The large distance of the Madeira archipelago to a continental source supports a human-mediated introduction of the house mice. Historical documents date the arrival of Portuguese settlers in the 1490 s pinpointing Portugal as a likely origin of the mouse colonists. This scenario is in agreement with the allozyme data since the mice from Madeira and Portugal show the highest levels of genic similarity. In addition, a Portuguese origin fits well with the observed level of diversity owing to the documented mode of colonization. After the discovery of Madeira, the island was rapidly settled to develop commercially lucrative crops such as sugar cane. Two main centres were established (Machico and Funchal; Albuquerque and Vieira, 1988) from which the colonization spread rapidly north, but mostly along the southern coast (Nunes et al., 2005). Thus, the colonization of Madeira was characterized by a large number of settlers and the rapid expansion of farm communities along the southern and eastern coastal areas. These features suggest that the initial mouse colonists were most likely not reduced in number, and that the developing human agricultural activities would have favoured their rapid post-founding expansion and dispersal. Such a colonization pattern not only would have considerably limited any loss in genetic variability (Zenger et al., 2003; Nei, 2005), but may even have contributed to its enhancement since the human immigrants originated from several regions in Portugal (Albuquerque and Vieira, 1988). Similarly, the lower genic diversity measured in the Porto Santo mice compared to those in Madeira is in agreement with records of a less intensive settlement both in population number and through time, suggesting fewer migration events and subsequent expansion. In Madeira, however, one would expect the initial genic diversity to have been subsequently eroded during the chromosomal radiation that took place in this island. Indeed, the extensive and rapid karyotypic divergence argues in favour of the successive fixation of Rb fusions through recurrent bottlenecks. Once again, the results show no traces of a reduction in genic diversity within the races related to processes of chromosomal differentiation. A previous study of mainland European Rb races investigated the values of bottleneck sizes, mutation rates and selective disadvantage coefficients compatible with a genic and chromosomal architecture similar to that found in the Madeira mice (Britton-Davidian et al., 1989). The computations yielded a realistic range of values sufficient to ensure a fixation rate in large-sized bottlenecks (>35 founding individuals) comparable to that observed. However, fixation rates in Madeira are higher than previous estimates by a factor of 10 (4.5 × 10−3 per generation vs 4 × 10−4 per generation), leading to the suggestion that additional genomic factors may be involved in the Rb process in this island. Two possibilities have been proposed, one of which consisted in elevated mutation rates such as those observed in laboratory strains (10−4 per generation), particularly those that have already fixed one Rb fusion (10−2 to 10−3 per generation; see Britton-Davidian et al., 2005). The other resides in a selective transmission advantage of Rb fusions in heterozygous females; such distortions have been documented in humans, but have yet to be demonstrated in house mice with a predominantly Rb karyotype (Pardo-Manuel de Villena and Sapienza, 2001).
A Portuguese origin of mice from Madeira contrasts with two main results of a previous mtDNA analysis. In effect, the study by Gündüz et al. (2001) clearly shows the presence in the Madeira mice of mtDNA haplotypes, which are characteristic of northwestern continental Europe. Similarly, the haplotypes present in the mice from Porto Santo are unique to this island and very closely related to the most frequent one in Madeira, pointing to a single and similar northern origin of the mice from these two islands. These results led the authors to postulate an earlier introduction by Danish Viking seafarers involving an accidental shipwreck on Madeira, as no trace of a prolonged presence is recorded. The allozyme data on the other hand favour a Portuguese origin for the Madeira mice and a separate one for the Porto Santo ones that are tentatively ascribed to Scotland. Thus, the analysis of allozyme and mtDNA markers indicate not only that the nuclear and mitochondrial genomes of the island mice have different founder origins, but that the geographic source populations of the nuclear component is not the same in Madeira (Portugal) and Porto Santo (Scotland). One possible explanation is compatible with an initial Viking introduction of mice as suggested by Gündüz et al. (2001), followed by a second one from Portugal and Scotland. Subsequent asymmetrical reproduction between these distinct sets of mice would have resulted in a maternal Viking signature with a nuclear Portuguese/Scottish genome. A trace of such a mixed background may explain the position of the Pod race, which clusters with the populations from Denmark in some of the analyses. Such asymmetric introgression is known to occur in several species and is related to differences in sex dispersal and/or social characteristics (Jones et al., 1995; Waits et al., 2000; Shaw, 2002). The fact that in both islands the original mitochondrial genome prevailed over those of the subsequent invaders suggests that selective processes may also be involved such as sexual or natural selection (Ryan and Wagner, 1987; Melo-Ferreira et al., 2005). The analysis of additional mitochondrial and nuclear markers on a larger sample of mice from different European localities would help clarify the historical and biological processes leading to the composite genome of the Madeira house mice.
Genic signature of chromosomal divergence
The role of chromosomal divergence in generating reproductive isolation can be measured by estimating levels of genic differentiation. In Madeira, however, it is difficult to tease apart the contribution of the chromosomal vs geographic barriers in limiting gene flow between races, as they are often coincident. This is the situation between the eastern races (San, Pod, Svi) and between these and the western ones (Edc, Adc, Ldb), which are the most chromosomally and genically differentiated and are isolated by geographic barriers. Several indications suggest in fact that geographic isolation may have been a determinant factor in shaping the differentiation in genic architecture. This is supported by the absence of a correlation between chromosomal and allozyme distances, although the karyotypic index used does not reflect precise chromosomal differences important in determining levels of underdominance and thus the strength of the barrier to gene flow. A case in point is Pod in which four of the six Rb fusions are unique to this race and which is also the most genically divergent of the Madeira races. Both of these characteristics suggest that the ancestral population was likely geographically isolated in this valley early in the divergence process, allowing this race to acquire a genic and chromosomal distinctiveness. The genic differences between the three western races also support the role of geographic barriers. These three parapatric races carry one Rb fusion each differing by a one-arm homology from the others: Rb(6.7) in Edc vs Rb(7.15) in Adc and Rb(5.18) in the latter vs Rb(15.18) in Ldb (see Figure 1); thus the same level of underdominance can be assumed in hybrids between them. The Edc and Adc races interact along a contact zone within which two chromosomal hybrids were subsequently recorded. These races are not genically differentiated, suggesting that the chromosomal differences are insufficient to impede gene flow. These results contrast with the significant genic differentiation between Edc and Ldb, which are separated by a short distance (Nunes et al., 2005). Thus, the general pattern of genic divergence seems to follow more closely that of geographic barriers than of chromosomal differentiation. However, this no longer holds true at the within-race scale, since some populations are clearly geographically distant (for example, Edc1 and Edc2) and show no significant genic differentiation. In these cases, the absence of a chromosomal barrier coupled with a large enough migration rate may have led to the genic cohesiveness of these populations. Such results will need to be reassessed with more variable markers, as the genic similarity may be due to a lack of resolution associated with the small sample size of the populations and the resilience of allozymes. The apparently low chromosomal impact on the genic architecture may be related to the time since the onset of the karyotypic divergence, and the rapid build-up in strength of the chromosomal barrier to gene flow as incompatible Rb fusions accumulate. So, a significant limitation in genic exchanges between races is only expected to occur at the later stages of chromosomal differentiation, and to affect more strongly genes located on incompatible fusions due to changes in recombination patterns (Dumas and Britton-Davidian, 2002). The specific signature ascribed to chromosomal rearrangements will need to be assessed by measuring the differential in gene flow of genes located on similar and different Rb fusions (Ayala and Coluzzi, 2005; Gavrilets and Vose, 2005). The Madeira races are ideally suited for such a study as they are derived from the same original gene pool, and have diverged parapatrically only very recently.
References
Albuquerque L, Vieira A (1988). The Archipelago of Madeira in the XV-Century Colecção Atlântica. Secretaria Regional do Turismo e Cultura & Centro de Estudos de História do Atlântico (eds). Região Autónoma da Madeira, Madeira, 69 pp.
Ayala FJ, Coluzzi M (2005). Chromosome speciation: humans, Drosophila and mosquitoes. Proc Natl Acad Sci 102: 6535–6542.
Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F (1996–2004). GENETIX 4.05, logiciel sous Windows TM pour la génétique des populations. Laboratoire Génome, Populations, Interactions, CNRS UMR. 5171, Université de Montpellier II: Montpellier, France.
Berry RL, Scriven PN (2005). The house mouse: a model and motor for evolutionary understanding. Biol J Linn Soc 84: 335–347.
Britton-Davidian J (1990). Genic differentiation in M. m. domesticus populations from Europe, the Middle East and North Africa: geographic patterns and colonization events. Biol J Linn Soc 41: 27–45.
Britton-Davidian J, Catalan J, Ramalhinho MG, Auffray J-C, Nunes AC, Gazave E, et al. (2005). Chromosomal phylogeny of Robertsonian races of the house mouse on the island of Madeira: testing between alternative mutational processes. Genet Res 86: 171–183.
Britton-Davidian J, Catalan J, Ramalhinho MG, Ganem G, Auffray J-C, Capela R et al. (2000). Rapid chromosomal evolution in island mice. Nature 403: 158.
Britton-Davidian J, Nadeau JH, Croset H, Thaler L (1989). Genic differentiation and origin of Robertsonian populations of the house mouse (Mus musculus domesticus Rutty). Genet Res 53: 29–44.
Corander J, Marttinen P, Mäntyniemi S (2006). Bayesian modification of stock mixtures from molecular data. Fish Bull 104: 550–558.
Dumas D, Britton-Davidian J (2002). Chromosomal rearrangements and evolution of recombination: comparison of chiasma distribution patterns in standard and Robertsonian populations of the house mouse. Genetics 162: 1355–1366.
Dupanloup I, Schneider S, Excoffier L (2002). A simulated annealing approach to define the genetic structure of populations. Mol Ecol 11: 2571–2581.
Emerson BC (2002). Evolution on oceanic islands: molecular phylogenetic approaches to understanding pattern and process. Mol Ecol 11: 951–966.
Emerson BC, Kolm N (2005). Species diversity can drive speciation. Nature 434: 1015–1017.
Excoffier L, Smouse PE, Quattro JM (1992). Analysis of molecular variance inferred from metric distances among haplotypes: application to human mitochondrial DNA restriction data. Genetics 31: 479–491.
Felsenstein J (1985). Phylogenies and the comparative method. Am Nat 125: 1–15.
Frankham R (1997). Do island populations have less genetic variation than mainland populations? Heredity 78: 311–327.
Gazave E, Catalan J, Ramalhinho MG, Mathias ML, Nunes AC, Dumas D et al. (2003). The non-random occurrence of Robertsonian fusion in the house mouse. Genet Res 81: 33–42.
Gavrilets S, Vose A (2005). Dynamic patterns of adaptative radiation. Proc Natl Acad Sci 102: 18040–18045.
Günduz I, Auffray J-C, Britton-Davidian J, Catalan J, Ganem G, Ramalhinho MG et al. (2001). Molecular studies on the colonization of the Madeiran archipelago by house mice. Mol Ecol 10: 2023–2029.
Jones CS, Noble LR, Jones JS, Tegelström H, Triggs GS, Berry RJ (1995). Differential male genetic success determines gene flow in an experimentally manipulated mouse population. Proc R Soc Lond B 260: 251–256.
Jordal BH, Hewitt GM (2004). The origin and radiation of Macaronesian beetles breeding in Euphorbia: the relative importance of multiple data partitions and population sampling. Syst Biol 53: 711–734.
Manel S, Schwartz MK, Luikart G, Taberlet P (2003). Landscape genetics: combining landscape ecology and population genetics. Trends Ecol Evol 18: 189–197.
Mathias ML, Nunes AC, Marques CC, Sousa I, Ramalhinho MG, Auffray JC et al. (2004). Adaptive energetics in house mice, Mus musculus L. from the island of Porto Santo (Madeira archipelago, North Atlantic). Comp Biochem Physiol A 137: 703–709.
Melo-Ferreira J, Boursot P, Suchentrunk F, Ferrand N, Alves PC (2005). Invasion form the cold past: extensive introgression of mountain hare (Lepus timidus) mitochondrial DNA into three other hare species in northern Iberia. Mol Ecol 14: 2459–2464.
Nachman MW, Boyer SN, Searle JB, Aquadro CF (1994). Mitochondrial DNA variation and the evolution of Robertsonian chromosomal races of house mice Mus domesticus. Genetics 136: 1105–1120.
Nachman MW, Searle JB (1995). Why is the house mouse karyotype so variable? Trends Ecol Evol 10: 397–402.
Navajas y Navarro M, Britton-Davidian J (1989). Genetic structure of insular Mediterranean populations of the house mouse. Biol J Linn Soc 36: 377–390.
Nei M (1978). Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89: 583–590.
Nei M (2005). Bottlenecks, genetic polymorphism, and speciation. Genetics 170: 1–4.
Nunes AC, Britton-Davidian J, Catalan J, Ramalhinho MG, Capela R, Mathias ML et al. (2005). Influence of physical environmental characteristics and anthropogenic factors on the position and structure of a contact zone between chromosomal races of the house mouse on the island of Madeira (North Atlantic, Portugal). J Biogeogr 32: 2123–2134.
Page RDM (1996). TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12: 357–358.
Pardo-Manuel de Villena F, Sapienza C (2001). Female meiosis drives karyotypic evolution in mammals. Genetics 159: 1179–1189.
Piálek J, Hauffe HC, Searle JB (2005). Chromosomal variation in the house mouse: a review. Biol J Linn Soc 84: 535–563.
Raymond M, Rousset F (1995). Genepop (Version 1.2): population genetics software for exact tests and ecumenism. J Hered 86: 248–249.
Ryan MJ, Wagner Jr WE (1987). Asymmetries in mating preferences between species: female swordtails prefer heterospecific males. Science 236: 595–596.
Salgueiro P, Coelho MM, Palmeirim JM, Ruedi M (2004). Mitochondrial DNA variation and population structure of the island endemic Azorean bat (Nyctalus azoreum). Mol Ecol 13: 3357–3366.
Shaw KL (2002). Conflict between nuclear and mitochondrial DNA phylogenies of a recent species radiation: what mtDNA reveals and conceals about modes of speciation in Hawaiian crickets. Proc Natl Acad Sci 99: 16122–16127.
Silvertown J (2004). The ghost of competition past in the phylogeny of island endemic plants. J Ecol 92: 168–173.
Smadja C, Catalan J, Ganem G (2004). Strong premating divergence in a unimodal hybrid zone between two subspecies of the house mouse. J Evol Biol 17: 165–176.
Swofford DL, Selander RB (1981). BIOSYS-1. A Computer Program for the Analysis of Allelic Variation in Population Genetics and Biochemical Systematics Release 1.7. University of Illinois: Urbana, IL.
Waits L, Taberlet P, Swenson JE, Sandegren F, Franzén R (2000). Nuclear DNA microsatellite analysis of genetic diversity and gene flow in the Scandinavian brown bear (Ursus arctos). Mol Ecol 9: 421–431.
Zenger KR, Richardson BJ, Vachot-Griffin AM (2003). A rapid population expansion retains genetic diversity within European rabbits in Australia. Mol Ecol 12: 789–794.
Acknowledgements
We are extremely grateful to Claude Berger who performed part of the allozyme analysis and to Ana Isabel Galvão, Carla Cristina Marques and Ruben Capela for their help in the field. We also thank Manuel Biscoito and Jorge Prudêncio. This study was financially supported by an ICCTI–French Embassy scientific collaboration, a grant from Fundação para a Ciência e a Tecnologia (PRAXIS/PCNA/C/BIA/135/96) and funding by a SAPIENS project (POCTI/BSE/47019/02). This is publication N°ISEM2007-062.
Author information
Authors and Affiliations
Corresponding author
Appendix A
Appendix A
The distribution of allelic frequencies at the 25 polymorphic loci in the samples studied is given in Appendix Table A1.
Rights and permissions
About this article
Cite this article
Britton-Davidian, J., Catalan, J., Lopez, J. et al. Patterns of genic diversity and structure in a species undergoing rapid chromosomal radiation: an allozyme analysis of house mice from the Madeira archipelago. Heredity 99, 432–442 (2007). https://doi.org/10.1038/sj.hdy.6801021
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6801021
Keywords
This article is cited by
-
Population structure and inbreeding in wild house mice (Mus musculus) at different geographic scales
Heredity (2022)
-
Inter-Population Genetic Variation in the Indian Pygmy Field Mouse Mus terricolor Blyth, 1851 (Rodentia, Muridae) from Terai and Dooars of Darjeeling Foothills
Proceedings of the Zoological Society (2019)
-
Meiotic behavior of a complex hexavalent in heterozygous mice for Robertsonian translocations: insights for synapsis dynamics
Chromosoma (2019)
-
Origin of the chromosomal radiation of Madeiran house mice: a microsatellite analysis of metacentric chromosomes
Heredity (2013)
-
Multiple origins of the western European house mouse in the Aeolian Archipelago: clues from mtDNA and chromosomes
Biological Invasions (2013)