Abstract
We studied phenotypic patterns and underlying quantitative genetics of development of sexual size dimorphism in red junglefowl (Gallus gallus). Using a multigenerational pedigree and the ‘animal model’ technique, we found significant heritability for many of the size and growth-related traits we examined, as well as significant genetic correlations among them. Despite sexual size dimorphism throughout posthatching ontogeny, the genetic correlation between males and females for all size measurements and growth parameters remained high. Significant positive phenotypic and genetic correlations between the fastest rate of growth and mass at week 26 (near asymptote) indicate that faster growth when young promotes larger adult size. However, age at which peak growth is reached does not appear to be phenotypically or genetically correlated with adult size. Positive genetic correlations within traits among ages were common, demonstrating that the genetic variance important to growth is relatively consistent among ages. However, male mass and tarsus length showed no genetic correlation between week 0 values and those from later ages. The body size traits of mass and tarsus length were genetically correlated with each other in females, but this pattern was not significant in males. Thus, despite striking sexual dimorphism in size and growth trajectories, size dimorphic traits in junglefowl show, with some exceptions, genetic integration between the sexes, among ages, and between traits.
Similar content being viewed by others
Introduction
The common form of sexual size dimorphism, males larger than females, appears to be the outcome of selection for success in male–male aggressive encounters (Ligon, 1999; Bennett and Owens, 2002). Behavioural ecologists interested in sexual selection have typically stopped their analyses at this level, and have ignored the developmental processes leading to sexual size dimorphism. However, there is a growing body of research devoted to understanding these mechanisms (Badyaev, 2002).
Functionally related morphological traits are typically genetically correlated with each other to a higher degree than are functionally unrelated traits (reviewed in Lynch and Walsh, 1998). Components of body size, for instance, often show high genetic correlation. Body size traits are presumably often under selection to covary phenotypically and thus be developmentally integrated (Cheverud, 1988), and the genetic correlations among these traits may be due to the influence of a partially overlapping set of genes (pleiotropy) on similar developmental pathways (Lynch and Walsh, 1998).
For sexually monomorphic traits, expression in both sexes is presumably influenced by the same developmental pathway and the same genetic variation, and thus genetic correlation between the sexes should be high (eg Jensen et al, 2003). With sexually size dimorphic traits, expectations for between-sex genetic correlations are less clear. Weakened genetic correlation might be expected during the evolutionary divergence between the sexes (Lande, 1980; Roff, 1997), although not necessarily (Badyaev, 2002). However, estimates of between-sex genetic correlations remain rare (Roff, 1997; Rice and Chippindale, 2001).
In this study, we assessed phenotypic variation in the development of two sexually size dimorphic traits. We then partitioned this phenotypic variation into genetic and environmental influences using multigenerational quantitative genetics techniques based on restricted maximum likelihood (REML) methods. These methods, developed by animal breeders, are becoming increasingly popular in behavioural ecology and evolutionary biology (reviewed in Kruuk, 2004). We used data from three generations of red junglefowl (Gallus gallus L.) to assess quantitative genetic patterns in mass gain and tarsus growth. We explored the extent to which growth trajectories were heritable and genetically integrated between traits and among ages. We also asked whether patterns of quantitative genetic variance were distinct in males and females and to what extent genetic correlations existed between the sexes.
Materials and methods
Study species
We studied patterns of growth in red junglefowl, the wild ancestor to the domestic chicken (see Parker and Garant, 2004 for details regarding our study population). Male dominance is an important determinant of reproductive access to females in this species (Collias and Collias, 1996), and males are much larger than females (Parker and Garant, 2004). Male mass and tarsus length both correlate positively with dominance rank (T Parker, unpublished data; Parker et al, 2002), probably because larger size bestows an advantage in male conflict.
In 6-month-old (nearly fully grown) junglefowl, mass and tarsus length are both highly heritable (Parker and Garant, 2004). However, these two traits showed different patterns of genetic variance and covariance in males and females. Mass and tarsus length were not genetically correlated in males, but were in females (Parker and Garant, 2004).
Data
Most of the birds used in these analyses were those included in previous work examining hypotheses related to sexual selection (Parker, 2003) as well as quantitative genetics of sexually dimorphic traits in a singe age class of junglefowl prior to their first breeding season (Parker and Garant, 2004). In brief, a founding generation of 17 males and 24 females of unknown relatedness hatched in 1998 were bred to produce a second generation in 1999. Of the 201 chicks from the 1999 generation measured over all or part of their growth period, 24 males and 56 females were bred to produce a third generation in 2000. Of the birds hatched in 2000, 321 were measured at least once during their development. Thus, 563 individuals are included in our pedigree. As some chicks died or were euthanized, and a few chicks were accidentally not measured at least once, sample sizes vary among ages. The smallest samples are for growth curve data, which required no missing measurements. This data set includes 205 males and 235 females. All birds produced in 1999 and 2000 were sired by artificial insemination. Females incubated their own eggs and reared chicks until 3 to 5 weeks old, at which point chicks were moved to standard density, single-age flocks containing up to 60 individuals. All birds were housed in large aviaries out of doors, were fed identical moderate protein poultry diets, and had ad libitum access to food and water. See Parker (2003) for additional details on the breeding and rearing conditions.
We did not take morphological measurements for birds in the founding population during their development, and so these birds only served to increase the number of links in the pedigree. In the two subsequent generations, we measured mass and tarsus length first at 2 days of age, then every 7 days until reaching the age of 10 weeks, and then every 4 weeks until 26 weeks of age. We measured mass to the nearest gram on an electronic balance and lengths with dial callipers to the nearest tenth of a millimetre. For each individual junglefowl for which we had no missing measurements (see above), we also fit Gaussian growth curves using a nonlinear optimization process (SAS 8.2, proc nlin) that determines the values of α, β and γ that minimize the sum of squares. We fit these curves for both mass and tarsus length to estimate the ages at which mass gain and tarsus growth rate were fastest and the steepness of the growth curves at those ages.
Data analysis
Identifying environmental correlates of morphology – We conducted general linear mixed-models analyses (SAS 8.2, proc mixed) to identify environmental variables correlated with the phenotypic traits of interest (tarsus length, mass, age at point of fastest growth, steepest slope of growth curve). This was done so that we could include these variables as fixed effects in our quantitative genetics analyses to account for heterogeneity in environmental effects on the phenotype. We included brood identity as a random variable in these mixed models to control for nonindependence of individuals within broods. The two variables found to be important influences on the morphological traits and growth were the individual's environment of rearing (three categories) and the individual's mother's environment of rearing (four categories, see Parker and Garant, 2004, for further explanation of these variables). The first was a consistent predictor of tarsus length, and so was selected for inclusion in further analyses of this trait, and the second was a consistent predictor of mass, and so was selected for inclusion in further analyses of this trait.
Heritability and components of variance – We estimated heritability of each trait through a mixed model REML estimation procedure using the software package ASReml (1.1, VSN International Ltd). We used the pedigree information (available for 563 individuals) to fit an individual ‘animal model’ (Lynch and Walsh, 1998), which partitions phenotypic variance in a quantitative character into its additive genetic and other fixed and random components (Kruuk, 2004). This technique can be used with unbalanced data sets and does not assume that selection, inbreeding, or assortative mating are absent (Kruuk, 2004). The animal model does assume a lack of selection in the founding generation, but it is relatively robust to violations of this assumption. The method also assumes that residual error from each observation is uncorrelated with other residual errors. This can be violated due to dominance genetic variance, but in practice, accounting for this problem is rarely accomplished (Lynch and Walsh, 1998). Trait values should be normally distributed, but the animal model is fairly robust to violations of this assumption (Kruuk, 2004). When we experimented with eliminating moderate outliers at the tails of our distributions, our estimates were not changed meaningfully. One of two categorical variables discussed above were included as fixed effects to account for temporal and spatial heterogeneity in environmental effects on the phenotype. The brood identity was always fitted as a random effect to account for common-environment effects specific to the individual brood, as well as maternal and paternal effects (Kruuk, 2004). Total phenotypic variance (VP) of each trait was therefore partitioned into additive genetic variance (VA), common environmental variance due to brood (VB) and residual variance (VR). The narrow-sense heritability (h2) was estimated as the ratio of the additive genetic variance (VA) to the total phenotypic variance (VP): h2=VA/VP. We assessed the significance of the additive genetic component of each model by comparing the full model with a reduced model lacking the additive genetic component using a likelihood ratio test (following a χ2 distribution, change in df between models=1) (see Wilson et al, 2005).
Genetic covariances – We used a multivariate animal model to calculate the genetic correlation among traits within and among sexes, and within and among ages. The same fixed and random effects used for heritability estimation were used in these analyses. We assessed genetic covariances in models containing two traits at a time (pairwise comparisons) because more complex models would not converge. We assessed genetic correlation among traits for which heritability estimates were not significant because large standard errors rather than small amounts of additive genetic variance explained the lack of significance for most traits. A meaningful role for additive genetic variance and covariance remained in these cases (the one exception being age of peak mass gain in males, for which there appeared to be almost no additive genetic variance, see Results). Genetic correlations were calculated as

Standard errors for heritabilities and genetic correlations were computed using the program ASReml. To determine whether including the genetic covariance significantly improved model fit, we used likelihood ratio tests to compare the models containing the genetic covariance component with reduced models in which the genetic covariance was fixed at 0.
Results
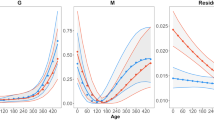
Phenotypic patterns of growth – Growth curves differed between the sexes, with males showing more rapid growth and a longer duration of growth than females (Figure 1). At 2 days after hatching, when the first measurements were taken, male junglefowl were not significantly heavier than females (following analyses conducted with SAS 8.2, proc mixed, including brood as random effect: F1,417=1.2, P=0.28; Table 1a) but had significantly longer tarsi (F1,417=28.4, P<0.0001; Table 1a). At all subsequent ages, males had highly significantly (F1,>339>301.3, P<0.0001; Table 1a) greater mass and longer tarsi than females. Females reached their fastest growth rate before males for both mass and tarsus length (F1,337>456.0, P<0.001; Figure 1, Table 1b). Although males had a faster mass gain (steeper slope of growth curve) at their peak than did females (F1,337=55.2, P<0.001; Figure 1a, Table 1b), the sexes did not differ in the steepest slope of their tarsus growth curves (F1,337=0.55, P=0.46; Figure 1b, Table 1b).
Parameter estimates for individual traits – For females, at all ages examined mass and tarsus length were heritable (Table 1a). This pattern was similar in males, although for tarsus at age 0 and 9 weeks, and mass at 9 weeks, the model containing an additive genetic component did not perform significantly better than a reduced model (Table 1a). For females, the two characteristics of growth curves we examined, age at fastest growth and steepest slope of the growth curve, were heritable for both tarsus growth and mass gain, although the additive genetic component did not significantly improve model fit for the age of fastest mass gain (Table 1b). For males, age at fastest mass gain showed almost no additive genetic variance, and thus no heritability (Table 1b). Although in males the other growth rate traits had heritability estimates >0.23, these were associated with large standard errors, and models including these additive genetic effects were not significantly better than reduced models without these effects (Table 1b).
Genetic correlations – Within traits, we found positive genetic correlations between the sexes for all models that converged (Table 2). Within each sex, genetic variance associated with mass or tarsus length was, for the most part, positively genetically correlated with the genetic variance influencing that trait at different ages (Table 3). However, in male chicks the genetic variance influencing mass and tarsus length at week 0 was not positively correlated with the genetic variance influencing these traits at later ages (Table 3). All other genetic correlations among ages were positive (Table 3). We observed positive genetic correlations between mass and tarsus length at almost all ages for both sexes, though in some cases, the model containing the genetic covariance component did not significantly outperform a reduced model (Table 4).
The steepest slope of the growth curve for mass was significantly positively genetically correlated with the week 26 mass for both males and females (Table 5). However, mass is not truly asymptotic at week 26, and so we cannot be sure growth rate genetically correlates with final mass. A similar, but weaker trend for tarsus length was unconvincing because of large standard errors and poor performance of the models containing the genetic covariance component in comparison with reduced models (Table 5). We did not assess genetic covariance involving the age at peak mass gain in males, because this trait appeared to have almost no additive genetic variance (Table 1). When we assessed age at peak mass gain in females, and the age of peak tarsus growth in males and females, we did not find good evidence of genetic correlations with week 26 mass or tarsus length, respectively (Table 5). Given the near total lack of a phenotypic correlation between these traits (Table 5), the lack of genetic correlation was likely.
We failed to get model convergence for the genetic correlation between growth parameters (age and slope of steepest growth) for female tarsus length (Table 5). For males, these growth parameters for tarsus length were negatively genetically correlated with each other (Table 5). However, the reduced model without the genetic covariance component did not converge, so we could not assess the relative effectiveness of the genetic covariance model. The negative genetic correlation indicates that genetic variance promoting an earlier peak tarsus growth rate also promotes a faster peak tarsus growth rate. We also observed a negative genetic correlation between age and slope of steepest growth for female mass, but this correlation was weaker, and the model containing the genetic covariance component did not significantly out perform the reduced model (Table 5).
Finally, in female junglefowl we observed positive genetic correlations between mass and tarsus length for both the age of peak growth and the slope of peak growth, although the model containing the genetic covariance significantly outperformed the reduced model only for slope (Table 5). In males, the genetic correlation between the slopes of mass and tarsus growth had large standard errors and the model containing the genetic covariance component did not outperform the reduced model (Table 5).
Discussion
For many trait measurements we examined, the variation among individuals had a significant genetic component. Throughout development, male and female mass and tarsus length, and their patterns of growth, at least in females, should be able to respond to selection. However, we also observed many significant genetic correlations among traits, between the sexes, and between the same traits at different ages, indicating that the trajectory of evolutionary responses of components of body size and growth traits to selection will be influenced by the selection acting on other correlated components of body size and growth (Roff, 1997).
Despite distinct sexual size dimorphism, we found significant genetic correlation between the sexes throughout development. This is consistent with our previous observation based on a larger sample of birds, but limited to measurements from individuals that were approximately 6 months old (Parker and Garant, 2004). Thus, the genetic variance responsible for mass gain and tarsus growth in males has a high degree of overlap with the genetic variance influencing mass gain and tarsus growth in females. A few of our estimates of genetic correlation between the sexes were greater than one. This may be due to relatively small sample sizes resulting in increased sampling variance. Although the precision of genetic correlation estimates may have been reduced by such effects, our conclusion that there are significant positive genetic correlations between male and female body size traits (and among traits within sex) is robust in light of the significantly improved fit of models containing the genetic covariance component in comparison with reduced models in which that covariance was fixed at one.
Genetic correlations in male chicks between measurements taken at week 0 and measurements taken later were all negative, although with very large standard errors indicating a lack of any real genetic correlation, negative or positive. This may appear inconsistent with the positive genetic correlations between week 0 male and female values because female traits were (where models converged) positively correlated with each other and with older male traits that were not positively correlated with male week 0 traits. However, the genetic correlation between male and female week 0 tarsus length did not significantly improve the fit of the model, and the genetic correlation between male and female week 0 mass could not be tested for significance because the reduced model did not converge. Also, because of large standard errors, some of these correlations are indistinguishable from moderate values, and so are not inconsistent with the apparent absence of positive genetic correlation between week 0 and later measurements in males. As just mentioned, in females we observed strong positive genetic correlations between week 0 measurements and measurements taken when older. This difference between males and females in the tendency for genetic correlation between early and later trait values may result from a strong influence of maternal genes on hatching size for both male and female chicks, as mediated through a genetic correlation between maternal mass and egg size (unpublished data). As found by Price and Grant (1985), recently hatched chicks resembled their mothers more than their fathers in body mass. In parent–offspring regressions (unpublished data), we found that mothers' mass at week 0 predicted both daughters' and sons' week 0 mass, but that fathers' week 0 mass did not predict the week 0 mass of either sons or daughters. However, the lack of genetic correlation between week 0 and later measurements for tarsus length in males does not appear consistent with this explanation. Neither maternal nor paternal week 0 tarsus lengths were good predictors of son or daughter week 0 tarsus lengths in parent–offspring regressions.
As we observed in red junglefowl, heavily selected lines of domestic chickens show high genetic correlations between the sexes, a decrease in maternal effects with age, and genetic correlations among growth curve traits (Mignon-Grasteau et al, 1999). Although artificial selection does not seem to have had major effects on quantitative genetic architecture, we do see phenotypic changes in ontogeny. Commercial domestic lines have body mass multiple times that found in junglefowl (Parker and Garant, 2004), and this is associated with a longer period of growth. However, the age at which peak growth rate is reached is also generally younger, often by several weeks, in domestic chickens than in junglefowl (Mignon-Grasteau et al, 1999). Thus, artificial selection has speeded up the onset of rapid growth and increased the duration of growth.
In conclusions, our analyses demonstrate that in red junglefowl, many size and growth measurements of sexually size dimorphic traits are not only heritable throughout ontogeny, but also generally are genetically correlated with each other. Much of the genetic variance in these traits appears to be part of a pool of genetic variance determining body size in both males and females. Our estimates of aspects of genetic variance and covariance are improved over those from traditional quantitative genetics analyses by our use of REML animal models (Kruuk, 2004). However, we still failed to acquire model convergence in some cases. Further studies using larger pedigree and alternative statistical methods not yet widely adopted by evolutionary biologists (eg random regression models; Meyer, 2004) should improve model convergence and thus broaden our understanding of genetic components of ontogeny.
References
Badyaev AV (2002). Growing apart: an ontogenetic perspective on the evolution of sexual size dimorphism. Trends Ecol Evol 17: 369–378.
Bennett PM, Owens IPF (2002). Evolutionary Ecology of Birds: Life Histories, Mating Systems, and Extinction. Oxford University Press: Oxford.
Cheverud JM (1988). The evolution of genetic correlation and developmental constraints. In: de Jong G (ed) Population Genetics and Evolution. Springer: Berlin.
Collias NE, Collias EC (1996). Social organization of a red junglefowl Gallus gallus population related to evolution theory. Anim Behav 51: 1337–1354.
Jensen H, Saether BE, Ringsby TH, Tufto J, Griffith SC, Ellegren H (2003). Sexual variation in heritability and genetic correlations of morphological traits in house sparrow (Passer domesticus). J Evol Biol 16: 1296–1307.
Kruuk LEB (2004). Estimating genetic parameters in natural populations using the ‘animal model’. Philos Trans Roy Soc London Ser B – Biol Sci 359: 873–890.
Lande R (1980). Sexual dimorphism, sexual selection, and adaptation in polygenic characters. Evolution 34: 292–305.
Ligon JD (1999). The Evolution of Avian Breeding Systems. Oxford University Press: Oxford.
Lynch M, Walsh B (1998). Genetics and Analysis of Quantitative Traits. Sinauer: Sunderland, MA.
Meyer K (2004). Scope for a random regression model in genetic evaluation of beef cattle for growth. Livest Prod Sci 86: 69–83.
Mignon-Grasteau S, Beaumont C, Le Bihan-Duval E, Poivey JP, de Rochambeau H, Ricard FH (1999). Genetic parameters of growth curve parameters in male and female chickens. Br Poultry Sci 40: 44–51.
Parker TH, Garant D (2004). Quantitative genetics of sexually dimorphic traits and capture of genetic variance by a sexually-selected condition-dependent ornament in red junglefowl (Gallus gallus). J Evol Biol 17: 1277–1285.
Parker TH, Knapp R, Rosenfield JA (2002). Social mediation of sexually selected ornamentation and steroid hormone levels in male junglefowl. Anim Behav 64: 291–298.
Parker TH (2003). Genetic benefits of mate choice separated from differential maternal investment in red junglefowl. Evolution 57: 2157–2165.
Price TD, Grant PR (1985). The evolution of ontogeny in Darwin's finches: a quantitative genetic approach. Am Nat 125: 169–188.
Rice WR, Chippindale AK (2001). Intersexual ontogenetic conflict. J Evol Biol 14: 685–693.
Roff DA (1997). Evolutionary Quantitative Genetics. Chapman & Hall: New York.
Wilson AJ, Coltman DW, Pemberton JM, Overall ADJ, Byrne KA, Kruuk LEB (2005). Maternal genetic effects set the potential for evolution in a free-living vertebrate population. J Evol Biol 18: 405–414.
Acknowledgements
We thank K Holsinger and an anonymous referee for constructive comments on the manuscript. JD Ligon provided advice throughout data gathering. The assistance of J Abbott, M Edgar, C Fellows, F Gurule, H Hill, J Mayfield, S Mercen, E O'Keefe, N Parker, G Quintana, R Ricci, Y Romero and J Rosenfield was essential during data collection. G Milliken assisted with growth curve calculations. While conducting these analyses, THP was supported by a US National Science Foundation (NSF) International Research Fellowship (INT-0202704) and DG was supported by a Natural Sciences and Engineering Research Council of Canada (NSERC) Postdoctoral Research Fellowship, and by a Biotechnology and Biological Sciences Research Council (BBSRC) grant to BC Sheldon and LEB Kruuk. Funding for data collection came from NSF (IBN-0072995), Sigma Xi, the American Ornithologists' Union, and various University of New Mexico sources. The research presented here was approved by the Institutional Animal Care and Use Committee of the Main Campus of the University of New Mexico.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Parker, T., Garant, D. Quantitative genetics of ontogeny of sexual dimorphism in red junglefowl (Gallus gallus). Heredity 95, 401–407 (2005). https://doi.org/10.1038/sj.hdy.6800742
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6800742
Keywords
This article is cited by
-
Genetic analysis of sexual size dimorphism in Markhoz goat
Tropical Animal Health and Production (2021)
-
Ontogeny of sex differences in the energetics and kinematics of terrestrial locomotion in leghorn chickens (Gallus gallus domesticus)
Scientific Reports (2016)
-
Correlated responses to selection on female egg size in male reproductive traits in a butterfly
Evolutionary Ecology (2009)