Abstract
Genetic diversity and genetic differentiation within and among island populations was examined by allozyme electrophoresis in Suzukia luchuensis (Labiatae), which is endemic to four of the Ryukyu Islands, southern Japan, and one island near Taiwan. Intrapopulation allozyme diversity was very low in all the four Ryukyu Islands, probably due to the effects of random drift in small populations. In contrast, genetic diversity at the species level was high, possibly because of an ancient origin of populations and/or multiple colonization of the species on different islands. Genetic differentiation among the overall populations was high (GST=0.863), while gene flow (Nm) as estimated from allozyme frequency data was 0.041, suggesting that its occurrence among populations is highly restricted. Hierarchical analysis of genetic differentiation indicated that a high proportion of the total allelic variance is attributed to variation among islands, corresponding to the fact that several alleles were fixed on only one island. However, intraisland genetic differentiation was small on all islands except Yonaguni Island, where S. luchuensis is relatively widely distributed. Most diversity was thus due to differences among islands.
Similar content being viewed by others
Introduction
Island floras are of great interest to evolutionary biologists (Stuessy and Ono, 1998). This is because they often contain many unique endemic species, which are morphologically divergent from continental relatives and are considered to have evolved in environments that differ from those of the mainland. The floras of some islands have an extremely high ratio of endemic species; for example, in Hawaii approximately 90% of the angiosperms are endemic (Groombridge, 1992). Often, gene flow from the mainland to an island is highly limited due to spatial isolation. This can result in rapid fixation of mutations and subsequent speciation (Barton, 1998). In addition, a lack of competition with other species and the possibility for colonization of new habitats may promote speciation on islands (Crawford et al, 1987).
Insular endemic plants are susceptible to extinction due to the negative impact of humans, competition and hybridization with alien plants, and the deterioration of their habitats (Rieseberg and Swenson, 1996; Maunder et al, 1998). They are thus of primary interest to conservation biologists. A major problem in this respect is that they often lack genetic variation (Frankham, 1997) and therefore will have difficulty in adapting to changing environments (Hamrick and Godt, 1995). Detailed examination of the genetic diversity of island floras has shown that insular endemic plants generally maintain a low level of genetic diversity and high genetic differentiation among populations (Francisco-Ortega et al, 2000; Crawford et al, 2001). However, levels of genetic diversity within species are substantially larger in endemics of the Canary Islands than in those of the Pacific islands, suggesting that the pattern and level of genetic diversity in endemic plants differs among different island groups (Crawford et al, 2001).
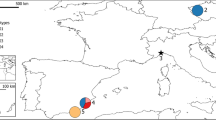
The Ryukyu Islands are located to the south of the Japan Archipelago and consist of ∼140 subtropical islands (Figure 1). Approximately 150 plant taxa, equivalent to 7% of the flora, are endemic to these islands (Hatusima, 1980), suggesting that the islands are not oceanic but continental in origin. For example, the proportion of endemics in the Ryukyu flora is comparable to that of several continental islands in the Mediterranean (see references in Affre and Thompson, 1997). In fact, previous geological studies showed that the islands have repeatedly had connections to the adjacent mainland (Kizaki and Oshiro, 1980). To date, genetic diversity has only been quantified in a small number of endemic plants in the Ryukyu Islands (Maki, 1999, 2001; Hiramatsu et al, 2001).
Distribution of the populations of S. luchuensis. The population codes and number of samples are shown in Table 1. Closed triangles indicate localities reported for S. luchuensis but not examined in this study.
Suzukia luchuensis Kudo (Labiatae) is a perennial herb occurring on rocky sites near the seashore. It creeps on the ground to reach a length of a few meters, and produces racemes near the top of stems. Its flowers are pink and 12–14 mm long. Pollinators for the species are not known. S. luchuensis has been reported to be distributed on only four islands of the Ryukyu Archipelago (Figure 1): Yonaguni Island, Kuroshima Island, Kume Island, and Okinawa Island and its contiguous small island (Okinawa Prefecture, 1996). It has also been recorded on Aragusuku Island (Chinen, 1993), but it is unknown whether it is still present here. Recently, the species was recorded on Lutao Island, east of Taiwan (Hsieh, 2000) (Figure 1). The species is listed in the national list of threatened plants (Red Data Book) in Japan as ‘vulnerable’ to extinction (Environment Agency of Japan, 2000), and the total number of extant individuals is estimated to be less than 1000. The species is subject to severe damage from trampling by humans (Environment Agency of Japan, 2000).
In this study, we address the following questions using enzyme electrophoresis. How much genetic diversity does S. luchuensis harbor at the species level and at the population level? Are levels of diversity larger or smaller than those of other insular endemic plants examined previously? To what extent does gene flow occur within and among island populations of S. luchuensis?
Materials and methods
Sampling populations
We sampled two populations of S. luchuensis from each of Kuroshima Island, Kume Island, and Okinawa Island and three populations from Yonaguni Island, where the species is more widely distributed (Figure 1). Population codes and the number of individuals sampled are given in Figure 1 and Table 1. Cuttings were collected individually from each population and samples were transported in plastic bags to the nursery at Tohoku University and planted in plastic pots filled with sand. Before electrophoresis, a sample of mature leaves was taken from each individual plant and transported on ice to the laboratory.
Allozyme electrophoresis
Enzyme extraction and polyacrylamide gel electrophoresis were carried out following the method of Maki and Murata (2001). The following 12 enzyme systems were examined by polyacrylamide gel electrophoresis: alanine aminopeptidase (AAP; EC 3.4.11.1), asparate amino transferase (AAT; EC 2.6.1.1), colometric esterase (EST; EC 3.1.1.–), formate dehydrogenase (FDH; EC 1.2.1.2), glutamate dehydrogenase (GDH; EC 2.7.1.1), glucose-6-phosphate isomerase (GPI; EC 5.3.1.9), NADP-dependent glyceraldehyde-3-phosphate dehydrogenase (G3PDH; EC 1.2.1.9), menazion reductase (MNR; EC 1.6.99.–), phosphoglucomutase (PGM; EC 5.4.2.2), 6-phosphogluconate dehydrogenase (6PGDH; EC 1.1.1.44), superoxide dismutase (SOD; EC 1.15.1.1), and triose-phosphate isomerase (TPI; EC 5.3.1.1). Staining protocols followed Tsumura et al (1990), except that for FDH and G3PDH, which followed Wendel and Weeden (1989).
Statistical analysis
Based on the results of allozyme electrophoresis, allele frequencies in each population of S. luchuensis were calculated for the loci encoding the 12 enzyme systems. The following indices were used to quantify the amount of genetic diversity within each population examined: the proportion of polymorphic loci (P) at the 95% criterion, the number of alleles per locus (A), and the expected heterozygosity (h). Genetic diversity parameters (P, A, and h) were also calculated at the species level. As in Hamrick and Godt (1989), we treated the loci polymorphic in at least one population as polymorphic at the species level.
Genetic differentiation among populations was estimated by gene diversity statistics corrected for errors due to small sample sizes (Nei and Chesser, 1983). The population genetic structure was analyzed by calculating GST values at each level of spatial hierarchy. GST was initially calculated among populations within an island using the genotype frequencies of each population. Following previous studies (Wright, 1978; Maki et al, 1996), we refer to this level as GPL, denoting differentiation among populations in an island. Since no enzyme polymorphism was found in Kuroshima Island, we regarded GPL for the island as zero for the following calculation. Next, the overall genotypic frequencies in each island enabled us to calculate a single GST, which is referred to as GLT. The amount of gene flow among populations was estimated as Nm=(1/GST−1)/4 (Slatkin and Barton, 1989).
Wright's (1951) fixation index (F) was estimated at each polymorphic locus as F=1−no/ne, where no is the observed number of heterozygotes and ne is the number of heterozygotes expected from the Hardy–Weinberg equilibrium. For loci with more than two alleles, the frequencies of the less-frequent alleles were combined into a single class. χ2 values for each polymorphic locus in a population were calculated as NF2, where N is the number of individuals per population (Li and Horvitz, 1953).
Standard genetic identity (I) (Nei, 1972), unbiased genetic identity (Î), and unbiased genetic distance (D̂) (Nei, 1978) were calculated for each pairwise comparison of all populations examined. A phenogram based on the unbiased genetic distance was obtained using the neighbor-joining method (Saitou and Nei, 1987) with PHYLIP version 3.5c (Felsenstein, 1993).
Results
Genetic diversity
A total of 20 putative loci were scored: Aap-1, Aap-2, Aat-1, Aat-2, Est-1, Est-2, Est-4, Fdh, Gdh, Gpi, G3pdh, Mnr-1, Mnr-2, 6pgdh, Pgm-1, Pgm-2, Sod-1, Sod-2, Tpi-1, and Tpi-2. In all populations, Fdh, Gpi, 6pgdh, Sod-2, Tpi-1, and Tpi-2 were monomorphic, and the other loci were polymorphic in at least one population.
Table 1 summarizes values of P, A, and h for each population and each island, and also at the species level. At the population level, the mean values of P, A, and h were 10.0, 1.13, and 0.031, respectively. The Kuroshima Island population contained no allozyme variation, while the Yonaguni Island population maintains the largest allozyme diversity.
The percentage of polymorphic loci (P=65.0) at the species level was over six times that of the mean population level (P=10.0), while gene diversity at the species level (h=0.232) was over seven times that of the gene diversity at the population level (h=0.031). The mean allele number per locus at the species level (A=1.90) was also higher than that at the population level (A=1.13) (Table 1).
Population genetic structure
The mean values of the fixation indices across the polymorphic loci for the populations ranged from −0.133 in population ON2 to 0.510 in population KM1 (Table 2). Approximately half of the fixation index (F) values (11 of 19) for each polymorphic locus deviated significantly from zero.
The total genetic diversity (HT) and the coefficient of gene differentiation (GST) overall for all populations were 0.232 and 0.863, respectively (Table 3). The latter value indicates that only 14% of the total genetic variation resides within a population, and the remaining 86% resides across populations. Gene flow among all populations per generation was extremely low (Nm=0.041). The GST value on Yonaguni Island (GST=0.440) suggests that genetic differentiation occurs among populations on this island. In contrast, little genetic differentiation occurs among populations on other islands. Consequently, gene flow among the populations per generation (Nm) was estimated to be larger in both Okinawa Island (Nm=4.852) and Kumejima Island (Nm=4.897) than in Yonaguni Island (Nm=0.318).
The results of hierarchical G-statistics showed that GPL and GLT are 0.134 and 0.848, respectively. This indicates that population differentiation within islands is low compared to that among islands. Indeed, with the exception of Yonaguni Isand, values of standard genetic identity (I) and unbiased genetic identity (Î) indicate that populations on the same island are not genetically differentiated, whereas genetic differentiation among islands is high (Table 4). The standard genetic identity (I) and the unbiased genetic identity (Î) for pairwise comparisons of the island populations are 0.566–0.837 and 0.529–0.777, respectively. Each island has at least one unique fixed allele not found on others (Figure 2). The Kume Island population has two fixed unique alleles at Aap-2 and Aat-1. In a neighbor-joining phenogram (Saitou and Nei, 1987) based on Nei's (1978) unbiased genetic distance, populations from the same island clustered together (Figure 3).
Phenogram for studied populations of S. luchuensis using the neighbor-joining method (Saitou and Nei, 1987) and Nei's (1978) unbiased genetic distance (D̂).
Discussion
Genetic diversity of S. luchuensis at the species level
The level of total allozyme genetic diversity in S. luchuensis (HT=0.232) is higher than has been estimated for many other plants endemic to oceanic islands. Crawford et al (2001) reported a mean total genetic diversity of 0.065 for 29 endemic plants in the Juan Fernandez Archipelago. Only one of the 29 species, Robisonia gracilis, contained a higher level of total genetic diversity than S. luchuensis. The mean total genetic diversity of 69 endemic plant taxa to the Pacific Islands is also very low (HT=0.064) (de Joode and Wendel, 1992). In contrast, Francisco-Ortega et al (2000) reported higher values of total genetic diversity for 69 Canary Island endemics (HT=0.186, range=0.047–0.456). However, only nine of these taxa had greater genetic diversity than S. luchuensis.
In contrast to oceanic islands such as the Juan Fernandez and Canary Islands, continental islands have a smaller proportion of endemic species. Genetic variation has been examined in only a few of these species, such as Cyclamen creticum, endemic to Crete and Karpathos in the Mediterranean, which contains a high total allozyme diversity (HT=0.216 for all nine loci examined) (Affre and Thompson, 1997). This value is comparable to that of S. luchuensis (HT=0.232). Other examples such as Hosta venusta endemic to Cheju Island, Korea (HT=0.315), and H. tsushimensis endemic to Tsushima Island, Japan (HT=0.207) (Chung, 1995), provide similar results. On the Ryukyu Islands, three other endemic plant species have been examined, yielding HT values of 0.134 in Aster asa-grayi (Maki, 1999), 0.188 in Lilium longiflorum (Hiramatsu et al, 2001), and 0.148 in Aster miyagii (Maki, 2001). Genetic diversity at the species level in S. luchuensis is thus more similar to those of plant species endemic to continental islands than those endemic to oceanic islands.
Crawford et al (2001) have discussed the reasons for the reduced level of genetic diversity of plants endemic to the Juan Fernandez Islands relative to those endemic to the Canary Islands. These authors suggest that Canary Island endemics are older lineages and therefore have accumulated many mutations since their origin. The Ryukyu Island endemics are also likely to be composed of older lineages, as the islands have a long geographic history of more than one period of landbridge connection with adjacent land masses. In the early Pleistocene (1.5–2.0 MYA), a long landbridge from central Ryukyu to Taiwan had been formed (Kizaki and Oshiro, 1980), although during the late Pleistocene (0.4–1.0 MYA) the sea level was 200 m higher than that at present, and lower islands were submerged and pushed upward thereafter (Kimura, 1996). However, many high-elevation islands have not been submerged since the Oligocene (Kizaki and Oshiro, 1980). The ancestor of S. luchuensis may have moved to the Ryukyu Archipelago during the early Pleistocene across the landbridge from Taiwan, and populations became isolated due to rising of sea level in the late Pleistocene. In S. luchuensis, the fact that each island population has at least one unique fixed allele that was not found in other island populations supports the older lineage hypothesis (Figure 2).
An alternative hypothesis to explain differences in genetic diversity of endemic plants between island groups is that multiple introductions from an adjacent continent are common for some island groups but not others (Crawford et al, 2001). Multiple introductions may be frequent in the Ryukyu Archipelago. For example, a close relative of S. luchuensis, namely S. shikikunensis, occurs in Taiwan, which is located in the southwest of Yonaguni Island. S. shikikunensis or its ancestral species may have colonized the Ryukyu Archipelago multiple times. However, it is unlikely that repeated independent speciation events have occurred on separate islands. This hypothesis thus has little support.
Genetic diversity within and among populations of S. luchuensis
Mean allozymic diversity within populations of S. luchuensis (HS=0.031) is comparable to the mean values recorded previously for insular endemic plants (HS=0.044 for 29 endemic species on the Juan Fernandez Islands, Crawford et al, 2001). Barrett (1996) found that genetic variation at allozyme loci is often reduced in island populations compared to those of mainland populations of the same or related species. The fact that insular endemic plants generally maintain low levels of within-population genetic diversity is probably due to random drift in small populations following a genetic bottleneck at the time of colonization. Except on the Yonaguhi Island, most populations of S. luchuenis are small and often consist of less than 50 individuals, and therefore may have been severely affected by random drift. A genetic bottleneck at the time of population establishment may have contributed to a low genetic diversity within populations; each island population of S. luchuensis would appear considered to have established sufficiently long ago in addition to allowing fixation of a unique allele on each island (Figure 2). A comparative genetic study of S. luchuensis and its Taiwanese relative S. shikikunensis would provide interesting data on this context.
Interpopulation genetic differentiation was found to be low within islands but high among islands. Compared to the findings presented in the previous reviews on insular endemic plants (Francisco-Ortega et al, 2000; Crawford et al, 2001), interpopulation genetic differentiation is extremely high in S. luchuensis (GST=0.863). In general, genetic differentiation among populations is larger in selfing species than outcrossing species (Hamrick and Godt, 1989). Fixation indices suggested that S. luchuensis is mainly a mixed mating or selfing species.
In addition to the mating system, interisland geographic isolation probably plays a major role in causing the extremely high genetic differentiation among the islands, as the hierarchical analysis of population genetic stucure in S. luchuensis (GLT=0.848) would suggest. Although pollinators of S. luchuensis are unknown at present, it seems unlikely that pollinators move between the islands, because the islands are isolated by long distances and populations are small and few. Accordingly, gene flow via pollen is less likely between islands. Since S. luchuensis is not found on any adjacent islands, long-distance migration via seeds is also likely to be a very rare event. Seeds of S. luchuensis do not have mechanisms specialized for wind dispersal such as wings, but may migrate by ocean currents. The genetic identities between pairs of island populations of S. luchuensis are nevertheless larger than most of those estimated for species pairs (Table 4) (Crawford, 1990). A study of ecological and morphological differentiation among island populations of S. luchuensis would help to obtain a better understanding of the ecology and evolution of the species in this archipelago.
References
Affre L, Thompson JD (1997). Population genetic structure and levels of inbreeding depression in the Mediterranean island endemic Cyclamen creticum (Primulaceae). Biol J Linn Soc 60: 527–549.
Barrett SCH (1996). The reproductive biology and genetics of island plants. Phil Trans R Soc Lond B 351: 725–733.
Barton NH (1998). Natural selection and random genetic drift as causes of evolution on islands. In: Grant P (ed) Evolution on Islands, Oxford University Press: Oxford. pp 102–173.
Chinen M (1993). Flora of the Sekisei Lagoon Islets, the Yaeyama Island. Island Stud Okinawa 11: 75–105 (in Japanese).
Chung MG (1995). Genetic diversity in two island endemics, Hosta venusta and H. tsushimensis (Liliaceae). J Jpn Bot 70: 322–327.
Crawford DJ (1990). Plant Molecular Systematics. Wiley Interscience: New York.
Crawford DJ, Ruiz E, Stuessy TF, Tepe E, Aqeveque P, Gonzalez F et al (2001). Allozyme diversity in endemic flowering plant species of the Juan Fernandez Archipelago, Chile: ecological and historical factors with implications for conservation. Am J Bot 88: 2195–2203.
Crawford DJ, Witkus R, Stuessy TF (1987). Plant evolution and speciation on oceanic islands. In: Urbanska KM (ed) Differentiation and Diversification of Land Plants, Springer-Verlag: Tokyo. pp 183–199.
de Joode DR, Wendel JF (1992). Genetic diversity and origin of the Hawaiian Island cotton, Gossypium tomentosum. Am J Bot 79: 1311–1319.
Environment Agency of Japan (2000). Threatened Wildlife of Japan. Red Data Book, 2nd edn. Vascular Plants. Japan Wildlife Research Center: Tokyo, Vol 8.
Felsenstein J (1993). PHYLIP (Phylogeny Inference Program) version 3.5c. Distributed by the author. Department of Genetics, University of Washington, Seattle, Washington, USA.
Francisco-Ortega J, Santos-Guerra A, Kim SC, Crawford DJ (2000). Plant genetic diversity in the Canary Islands: a conservation perspective. Am J Bot 87: 909–919.
Frankham R (1997). Do islands population have less genetic variation than mainland populations? Heredity 78: 311–327.
Groombridge B . (ed) (1992). Global Biodiversity: Status of the Earth's Living Resources. Chapman & Hall: London.
Hamrick JL, Godt MJW (1989). Allozyme diversity in plant species. In: Brown AHD, Clegg MT, Kahler AL, Wier BS (eds) Plant Population Genetics, Sinauer: Sunderland. pp 43–63.
Hamrick JL, Godt MJW (1995). Conservation genetics of endemic plant species. In: Avise JC, Holsinger KE (eds) Conservation Genetics, Chapman & Hall: New York. pp 281–301.
Hatusima S (1980). Origin of the flora in the Ryukyu Islands. In: Kizaki K (ed) Natural History of the Ryukyus, Tsukiji-shokan: Tokyo. pp 113–123 (in Japanese).
Hiramatsu M, II K, Okubo H, Huang KL, Huang CW (2001). Biogeography and origin of Lilium longiflorum and L. formosanum (Liliaceae) endemic to the Ryukyu Archipelago and Taiwan as determined by allozyme diversity. Am J Bot 88: 1230–1239.
Hsieh TH (2000). Suzukia Kud (Lamiaceae) in Taiwan. Taiwania 45: 217–225.
Kimura M (1996). Quaternary paleogeography of the Ryukyu Arc. J Geogr 105: 259–295 (in Japanese with English summary).
Kizaki K, Oshiro I (1980). The origin of the Ryukyu Islands. In: Kizaki K (ed) Natural History of the Ryukyus, Tsukiji-shokan: Tokyo. pp 8–37 (in Japanese).
Li CC, Horvitz DG (1953). Some methods of estimating the inbreeding coefficient. Am J Human Genet 5: 107–117.
Maki M (1999). Genetic diversity in the threatened insular endemic plant Aster asa-grayi (Asteraceae). Plant Syst Evol 217: 1–9.
Maki M (2001). Genetic differentiation within and among island populations of the endangered plant Aster miyagii (Asteraceae), an endemic to the Ryukyu Islands. Am J Bot 88: 2189–2194.
Maki M, Masuda M, Inoue K (1996). Genetic diversity and hierarchical population structure of a rare autotetraploid plant, Aster kantoensis (Asteraceae). Am J Bot 83: 296–303.
Maki M, Murata J (2001). Allozyme analysis of the hybrid origin of Arisaema ehimense (Araceae). Heredity 86: 87–93.
Maunder M, Culham A, Hankamer C (1998). Picking up the pieces: botanical conservation on degraded oceanic islands. In: Fiedler PL, Kareiva PM (eds) Conservation Biology for the Coming Decade, 2nd edn, Chapman & Hall: New York. pp 317–344.
Nei M (1972). Genetic distance between populations. Am Nat 106: 283–292.
Nei M (1978). Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89: 583–590.
Nei M, Chesser RK (1983). Estimation of fixation indices and gene diversities. Ann Hum Genet 47: 253–259.
Okinawa Prefecture (1996). Red Data Book of Okinawa Prefecture. Okinawa Prefecture: Naha.
Rieseberg LH, Swenson SM (1996). Conservation genetics of endangered island plants. In: Avise JC, Holsinger KE (eds) Conservation Genetics, Chapman & Hall: New York. pp 305–334.
Saitou N, Nei M (1987). The neighbor joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425.
Slatkin M, Barton NH (1989). A comparison of three indirect methods for estimating average levels of gene flow. Evolution 43: 1349–1368.
Stuessy TF, Ono M (eds) (1998). Evolution and Speciation of Island Plants. Cambridge University Press: Cambridge.
Tsumura Y, Tomaru N, Suyama N, Na'eim M, Ohba K (1990). Laboratory manual of isozyme analysis. Bull Tsukuba Univ Forests 6: 63–95 (in Japanese).
Wendel JF, Weeden NF (1989). Visualization and interpretation of plant isozymes. In: Soltis DE, Soltis PS (eds) Isozymes in Plant Biology, Dioscorides Press: Portland. pp 5–45.
Wright S (1951). The genetical structure of populations. Ann Eugen 15: 323–354.
Wright S (1978). Evolution and the genetics of populations. Vol. 4, Variability Within and Among Natural Populations. University Chicago Press: Chicago.
Acknowledgements
The authors thank S Horie for sampling and K Sato for the cultivation of S. luchuensis. Thanks are also due to J Yokoyama and T Fukuda for their comments.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Maki, M., Yamashiro, T. & Matsumura, S. High levels of genetic diversity in island populations of the island endemic Suzukia luchuensis (Labiatae). Heredity 91, 300–306 (2003). https://doi.org/10.1038/sj.hdy.6800311
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6800311
Keywords
This article is cited by
-
Conservation genetics and potential geographic distribution modeling of Corybas taliensis, a small ‘sky Island’ orchid species in China
BMC Plant Biology (2024)
-
Conservation genetics and genetic mating system of the yellow-shouldered blackbird (Agelaius xanthomus), an endangered island endemic
Conservation Genetics (2015)
-
Temperate origin and diversification via southward colonization in Fatsia (Araliaceae), an insular endemic genus of the West Pacific Rim
Tree Genetics & Genomes (2014)
-
Genetics, Evolution, and Conservation of Island Plants
Journal of Plant Biology (2010)
-
Isolation and characterization of 12 microsatellite loci from Suzukia shikikunensis (Lamiaceae), a genus endemic to Taiwan and Ryukyus
Conservation Genetics (2008)