Abstract
Purpose
To investigate the transforming growth factor beta-induced gene (TGFBI) mutations in Brazilian patients with corneal dystrophy and to evaluate the phenotype–genotype correlation in these patients.
Methods
A total of 11 unrelated families were studied. The diagnosis of corneal dystrophy was based on clinical and histopathological findings. Genomic DNA was extracted from peripheral blood leucocytes, and exons 4 and 12 of the TGFBIgene were amplified by polymerase chain reaction followed by direct sequencing on both strands.
Results
Five different mutations in the TGFBIgene were found in the probands. We identified the following mutations: lattice corneal dystrophy—R124C and A546T; Reis-Bücklers corneal dystrophy—R555Q and R124L; granular corneal dystrophy—R555W and Avellino dystrophy—R555W. In three of the 11 studied families there was no mutation in exons 4 and 12.
Conclusions
This is the first report of mutations in the TGFBIgene in a series of Brazilian patients with corneal dystrophy. The findings indicate that TGFBIgene screening should be considered in the diagnosis of corneal dystrophy.
Similar content being viewed by others
Introduction
Corneal dystrophies are inherited disorders characterized by bilateral, symmetrical opacities. The corneal opacities are caused by progressive accumulation of deposits resulting in loss of transparency and visual deterioration.1, 2
Recent molecular analysis showed that different types of corneal dystrophies are caused by mutations in the transforming growth factor beta-induced gene (TGFBI), located on chromosome 5q31.2, 3, 4 Several phenotypes are caused by specific mutations in the TGFBI gene such as: lattice corneal dystrophy type I (CDL1, OMIM122200), lattice corneal dystrophy type IIIA (LCDIIIA), granular corneal dystrophy (CDGG1, OMIM 121900), Avellino corneal dystrophy (ACD, OMIM 121900), Reis-Bücklers's corneal dystrophy (CDRB, OMIM 121900), and Thiel-Behnke corneal dystrophy (CDB2, OMIM 602082).1, 4, 5
Keratoepithelin, the protein product of the TGFBI gene, is an extracellular matrix protein expressed in many tissues including corneal epithelium.4, 6 Most patients have mutations at mutational hot spots, corresponding to arginine 124 and 555 of the keratoepithelin protein.5, 6
The diagnosis of corneal dystrophy is usually based on the clinical findings and the histopathological characteristics of the corneal deposits. The genotypic approach contributes to the assessment and refinement of the diagnosis of corneal dystrophies particularly in the atypical cases.2, 4, 5
Mutations in the TGFBI gene have been described in patients from many nationalities.7, 8, 9, 10, 11, 12 We report the first series of Brazilian patients screened for mutations in the TGFBI gene.
Materials and methods
This study was approved by the ethics committee of the Federal University of São Paulo and conformed to the tenets of the Declaration of Helsinki. Informed consent from all participants was obtained for clinical and molecular genetic study. In all, 11 unrelated families with corneal dystrophy were studied, including patients with clinical diagnosis of lattice dystrophy type I (CDL1), Reis-Bücklers's dystrophy (CDRB), granular dystrophy (CDGG), and Avellino dystrophy (ACD). The diagnoses were confirmed by histopathology in eight patients after corneal transplantation.
Molecular analysis
DNA of the 11 probands was extracted from peripheral blood using standard procedures. Genomic DNA of exons 4 and 12 of the TGFBI gene were amplified using appropriate forward and reverse primers.9 Each polymerase chain reaction (PCR) was carried out in 50 μl reaction mixture containing DNA (200 ng), primers (0.4 μM each), MgCl2 (1.5 mM), dNTPs (0.2 mM), 1 × PCR buffer and Taq DNA polymerase (0.5 U). The PCR conditions were as follows: 10 min at 94°C, followed by 35 cycles of 1 min at 94°C, annealing temperature for 1 min, and 72°C for 1 min, with a final extension at 72°C for 5 min. The annealing temperature was 60°C for exon 4 and 55°C for exon 12. The PCR products were purified with the QIAquik PCR purification kit (Qiagen Inc., ONT, Canada) and sequenced on both strands using a dye terminator cycle sequencing kit (Amersahm Biosciences). The sequencing results were compared with the nucleotide sequence of the published TGFBI cDNA.13 Exons 11, 13, and 14 of the TGFBI gene were also investigated on the proband of family 1, diagnosed as lattice corneal dystrophy.
Results
In all, 45 Brazilian patients from 11 unrelated families with corneal dystrophy have been investigated. DNA samples from 11 probands affected with lattice corneal dystrophy (three families), Reis-Bücklers's corneal dystrophy (four families), granular corneal dystrophy (three families), and Avellino dystrophy (one family) were screened for mutations within the TGFBI gene. The two mutational hot spots (R124 and R555) were analysed by direct sequencing of exons 4 and 12.
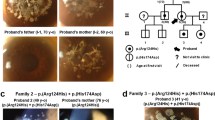
There were five different mutations in the 11 studied probands (Table 1). Sequencing analysis revealed two different mutations in patients with lattice corneal dystrophy (families 2 and 3). The proband of family 2 presented a base pair transition at nucleotide 417 (C → T), which converts an arginine into cysteine at codon 124 (R124C). The proband of family 3 presented the A546T (G → A) mutation at codon 546. There was no mutation in the hot spots of the proband of family 1. In this case exons 11, 13, and 14 were also investigated but no mutations were detected.
In the cases with diagnosis of Reis-Bücklers's dystrophy two different mutations were detected R555Q and R124L. Three probands (families 4, 5, and 6) presented a base pair transition at nucleotide 1711 (G → A), which converts an arginine into glutamine at codon 555 (R555Q) and one had the R124L mutation with a G → T transition at position 418.
In one of the three families with granular dystrophy we found the R555W mutation (Figure 1). There was no mutation in exons 4 and 12 in the two other families diagnosed with granular dystrophy. The proband of family 11 diagnosed as Avellino dystrophy presented the R555W mutation.
Discussion
We searched for mutations in the TGFBI gene in 11 unrelated Brazilian families affected with different types of corneal dystrophies. The sequencing analysis of both strands of exons 4 and 12 revealed mutations in eight families.
The lattice dystrophy cases presented two mutations, R124C and A546T, that were previously described by other authors.4, 5, 14 In family 1, there was no mutation in exons 4 and 12 of the TGFBI gene. This family had an unusual aspect in biomicroscopy, similar in all affected members, with peripheral corneal neovascularization and severe stromal opacity. The lattice diagnosis was based on histopathologic finding of amyloid stromal deposits in several affected members that were submitted to corneal transplant. In this case, mutation could be present within another exon of the TGFBI gene. Several studies reported mutations in exons 11, 13, and 14 of the TGFBI gene in cases of lattice dystrophy.1, 7, 15, 16 As the proband of family 1 have not shown mutation in exons 4 and 12 we investigated exons 11, 13, and 14 but the sequence analysis have not detected mutation. Many reasons can explain this result. The mutation in this case could be localized in other exon or intron of the TGFBI gene or could be detected at another gene.16, 17
The A546T mutation in the TGFBI gene that had been detected in the proband of family 2, was previously reported in cases of lattice dystrophy type IIIA-like that exhibited deposits similar to those observed in LCDI.14
In four probands diagnosed with Reis-Bücklers's corneal dystrophy, three were found to have the R555Q mutation reported to cause Thiel-Behnke dystrophy, whereas only one had the R124L mutation reported to cause Reis-Bücklers's dystrophy. Molecular biology findings had shown that most of the cases diagnosed as Reis-Bücklers's dystrophy are actually cases of Thiel-Behnke dystrophy. The differential diagnosis can be made by clinical and histological features or by the genetic findings.18, 19, 17 In our study, the families related to the R555Q mutation presented an early onset of subepithelial corneal opacities associated with recurrent erosions suggestive of Reis-Bücklers's dystrophy.
Although the proband of family 8 with granular dystrophy had the R555W mutation, two probands (families 9 and 10) with characteristic biomicroscopic findings did not present any mutation in exons 4 and 12 of the TGFBI gene. It is possible that mutation in another exon or in introns could be associated with these cases.
The proband clinically diagnosed as Avellino corneal dystrophy (family 11) presented a R555W mutation that is associated with granular dystrophy. It is well known that the initial manifestation in Avellino dystrophy presents only granular lesions demonstrating that these two conditions can be confused when the diagnosis is based on the clinical aspects especially in early cases.
Our results confirm the mutation hot spots at positions R124 and R555, as only one of the mutations identified in this study was located at other position (A456T). In four cases, DNA analysis altered the prior diagnosis based on clinical findings, including three families with initial diagnosis of Reis-Bückler's dystrophy (families 4, 5, and 6) and one family with diagnosis of Avellino dystrophy (family 11). These findings demonstrate that gene analysis may be useful in cases where the clinical features and the histopathological findings of the corneal dystrophy may not be conclusive.
All inherited corneal dystrophies caused by mutations in the TGFBI gene are associated with an extracellular deposition of protein within the cornea.15, 20, 21 Immunohistochemical studies demonstrated that the mutated keratoepithelin is a major component of the corneal deposits.20, 21, 22 The keratoepithelin protein has four internal repeat domains and most of the mutations reported are located in the amino acid R124 or in the fourth fascilin-like domain.
The mutations in the TGFBI gene cause a range of different types of opacities encompassing amyloid and nonamyloid deposits.4, 6 Several mechanisms have been implicated in the pathogenesis of the TGFBI-related corneal dystrophies. Some authors suggest that mutations could affect the tertiary structure of keratoepithelin causing the protein subunits to polymerize into different structures or that the modified domains could impair the binding of keratoepithelin to stromal proteins such as collagen type VI affecting the structural characteristics of the tissue.4, 6, 10, 15 Some studies indicate that mutations at amino acid R124 abolish a phosphorylation site that could affect the tertiary structure of keratoepithelin leading to amyloid formation.4, 20, 22, 23
Further studies are necessary to understand the role of keratoepithelin in normal corneal structure and function. At the moment knowledge about genotype/phenotype correlations and the mutation status can help with predictions of disease progression and recurrence after treatment.
In conclusion, this is the first report of TGFBI gene mutation in Brazilian patients with corneal dystrophies. Our results confirm the importance of TGFBI gene mutation analysis in the diagnosis of corneal dystrophies.
References
Scmitt-Bernard CF, Guittard C, Arnaud B, Demaille J, Argiles A, Claustres M et al. BIGH3 exon 14 mutations lead to intermediate type I/IIIA of lattice corneal dystrophies. Invest Ophthalmol Vis Sci 2000; 41: 1302–1308.
Stone EM, Mathers WD, Rosenwasser GOD, Holland EJ, Folberg R, Krachmer JH et al. Three autosomal dominant corneal dystrophies map to chromosome 5q. Nat Genet 1994; 6: 47–51.
Small KW, Mullen L, Barletta J, Graham K, Glasgow B, Stern G et al. Mapping of Reis-Bücklers’corneal dystrophy to chromosome 5q. Am J Ophthalmol 1996; 121: 384–390.
Munier FL, Korvatska E, Djemaï A, Le Paslier D, Zografos L, Pescia G et al. Kerato-epithelin mutations in four 5q31-linked corneal dystrophies. Nat Genet 1997; 15: 247–251.
Korvatska E, Munier FL, Djemaï A, Wang MX, Frueh B, Chiou AG-Y et al. Mutation hot spots in 5q31-linked corneal dystrophies. Am J Hum Genet 1998; 62: 320–324.
Chakravarthi SVVK, Kannabiran C, Sridhar MS, Vemuganti GK . TGFBI gene mutations causing lattice and granular corneal dystrophies in Indian patients. Invest Ophthalmol Vis Sci 2005; 46: 121–125.
Fujiki K, Hotta Y, Nakayasu K, Yamaguchi T, Kato T, Uesugi Y et al. Six different mutations of the TGFBI (βig-h3, keratoepithelin) gene found in Japanese corneal dystrophies. Cornea 2000; 19(6): 842–845.
Kim HS, Yoon SK, Cho BJ, Kim EK, Joo CK . BIGH3 gene mutations and rapid detection in Korean patients with corneal dystrophy. Cornea 2001; 20(8): 844–849.
Yoshida S, Kumano Y, Yoshida A, Hisatomi T, Matsui H, Nishida T et al. An analysis of BIGH3 mutations in patients with corneal dystrophies in the Kyushu district of Japan. Jpn J Ophthalmol 2002; 46: 469–471.
Pampukha VM, Drozhyna GI, Livshits LA . TGFBI gene mutation analysis in families with hereditary corneal dystrophies from Ukraine. Ophthalmologica 2004; 218: 411–414.
Chakravarthi SV, Kannabiran C, Sridhar MS, Vemuganti GK . TGFBI gene mutations causing lattice and granular corneal dystrophies in Indian patients. Invest Ophthalmol Vis Sci 2005; 46: 121–125.
Tian X, Fujiki K, Murakami A, Xie P, Kanai A, Liu Z . Novel mutation (V505D) of the TGFBI gene found in a Chinese family with lattice corneal dystrophy, type I. Jpn J Ophthalmoll 2005; 49: 84–88.
Skonier J, Neubauer M, Madisen L, Bennett K, Plowman GD, Purchio AF . cDNA cloning and sequence analysis of βig-h3, a novel gene induced in a human adenocarcinoma cell line after treatment with transforming growth factor-β. DNA Cell Biol 1992; 11(7): 511–522.
Dighiero P, Valleix S, D'Hermies F, Drunat S, Ellies P, Savoldelli M et al. Clinical, histologic, and ultrastructural features of the corneal dystrophy caused by the R124L mutation of the BIGH3 gene. Ophthalmology 2000; 107: 1353–1357.
Klintworth GK, Bao W, Afshari NA . Two mutations in the TGFBI (BIGH3) gene associated with lattice corneal dystrophy in an extensively studied family. Invest Ophthalmol Vis Sci 2004; 45: 1382–1388.
Warren JF, Abbott RL, Yoon MK, Crawford JB, Spencer WH, Margolis TP . A new mutation (Leu569Arg) within exon 13 of the TGFBI (BIGH3) gene causes lattice corneal dystrophy type I. Am J Ophthalmol 2004; 136(5): 872–878.
Ellies P, Renard G, Valleix S, Boelle PY, Dighiero P . Clinical outcome of eight BIGH3-linked corneal dystrophies. Ophthalmology 2002; 109: 793–797.
Küchle M, Green WR, Völcker HE, Barraquer J . Reevaluation of corneal dystrophies of Bowman's layer and the anterior stroma (Reis-Bücklers and Thiel-Behnke types): a light and electron microscopic study of eight corneas and a review of the literature. Cornea 1995; 14(4): 333–354.
Mashima Y, Nakamura Y, Noda K, Konish M, Yamada M, Kudoh J et al. A novel mutation at codon 124 (R124L) in the BIGH3 gene is associated with a superficial variant of granular corneal dystrophy. Arch Ophthalmol 1999; 117: 90–93.
Korvatska E, Munier FL, Chaubert P, Wang MX, Mashima Y, Yamada M et al. On the role of kerato-epithelin in the pathogenesis of 5q31-linked corneal dystrophies. Invest Ophthalmol Vis Sci 1999; 40: 2213–2219.
Streeten BW, Qi Y, Klintworth GK, Eagle RC, Strauss JA, Bennett K . Immunolocalization of the big-h3 protein in 5q31-linked corneal dystrophies and normal corneas. Arch Ophthalmol 1999; 117: 67–75.
Korvatska E, Henry H, Mashima Y, Yamada M, Bachmann C, Munier FL et al. Amyloid and non-amyloid forms of 5q31-linked corneal dystrophy resulting from kerato-epithelin mutations at Arg-124 are associated with abnormal turnover of the protein. J Biol Chem 2000; 275: 11465–11469.
Munier FL, Frueh BE, Othenin-Girard P, Uffer S, Cousin P, Wang MX et al. BIGH3 mutation spectrum in corneal dystrophies. Invest Ophthalmol Vis Sci 2002; 43: 949–954.
Author information
Authors and Affiliations
Corresponding author
Additional information
The authors do not have any commercial or proprietary interest in the products used in this study
Rights and permissions
About this article
Cite this article
Solari, H., Ventura, M., Perez, A. et al. TGFBI gene mutations in Brazilian patients with corneal dystrophy. Eye 21, 587–590 (2007). https://doi.org/10.1038/sj.eye.6702264
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.eye.6702264