Abstract
Background:
The RAS/RAF/MEK/ERK pathway is involved in the balance between melanocyte proliferation and differentiation. The same pathway is constitutively activated in cutaneous and uveal melanoma (UM) and related to tumour growth and survival. Whereas mutant BRAF and NRAS are responsible for the activation of the RAS/RAF/MEK/ERK pathway in most cutaneous melanoma, mutations in these genes are usually absent in UM.
Methods:
We set out to explore the RAS/RAF/MEK/ERK pathway and used mitogen-activated protein kinase profiling and tyrosine kinase arrays.
Results:
We identified Src as a kinase that is associated with ERK1/2 activation in UM. However, low Src levels and reduced ERK1/2 activation in metastatic cell lines suggest that proliferation in metastases can become independent of Src and RAS/RAF/MEK/ERK signalling. Inhibition of Src led to the growth reduction of primary UM cultures and cell lines, whereas metastatic cell line growth was only slightly reduced.
Conclusion:
We identified Src as an important kinase and a potential target for treatment in primary UM. Metastasis cell lines seemed largely resistant to Src inhibition and indicate that in metastases treatment, a different approach may be required.
Similar content being viewed by others
Main
Uveal melanoma (UM) is a rare neoplasm that arises from melanocytes in the eyes. It usually affects people in their sixties with an incidence rate of ∼6–8 new cases per million per year among Caucasians (Egan et al, 1988; Singh and Topham, 2003). Little is known about the molecular pathogenesis of UM compared with cutaneous melanoma (CM). Cutaneous melanoma and UM share the same embryonic origin and similar histological features, but mutations that regulate proliferation and cause a loss of cell-cycle control in CM can hardly be found in UM. Whereas p16-regulated cell cycle control is targeted by the deletion of chromosome 9p or by the mutation of CDKN2A in CM, most of the UM cell lines posses a wild-type p16-encoding gene that is, however, not expressed because of the epigenetic modification of the CDKN2A gene (van der Velden et al, 2001). The same may be true for the activation of the RAS-RAF-MEK-ERK or the classical mitogen-activated protein kinase (MAPK) pathway. The MAPK activation is crucial for the development of melanocytic neoplasia, and a constitutive activation of this pathway has been associated with many different types of cancer (Goding, 2000; Reddy et al, 2003). In CM, the activation of the MAPK pathway has been shown to occur by a variety of mechanisms, including autocrine growth factor stimulation and mutation of the NRAS (20% of cases) and BRAF (60% of cases) genes (van Elsas et al, 1995; Davies et al, 2002; Satyamoorthy et al, 2003). The BRAF mutations have only rarely been reported in UM and activating mutations in NRAS, which are found in 25% of all cancers, have never been reported (Mooy et al, 1991; Soparker et al, 1993; Cohen et al, 2003; Cruz III et al, 2003; Edmunds et al, 2003; Rimoldi et al, 2003). However, we and others have found that UM is heterogeneous and that, with more sensitive techniques, the percentage of mutant BRAF-positive UM may be higher (Janssen et al, 2008; Maat et al, 2008). The lack of mutations in the majority of cells is in contrast with immunohistochemistry and western blot analysis, which have shown activation of ERK1/2 in most UM (Rimoldi et al, 2003; Weber et al, 2003; Zuidervaart et al, 2005). Nevertheless, the pharmacological inhibition of MAPK/ERK kinases 1 and 2 (MEK1/2) and the genetic targeting of BRAF with siRNA resulted in a reduced proliferation of UM cell lines (Lefevre et al, 2004; Calipel et al, 2006). This indicates that although mutations are absent, the RAS/RAF/MEK/ERK pathway is essential for UM growth and suggests that an upstream factor is involved in autonomous UM proliferation. Recently, c-Kit was shown to be upregulated in UM and involved in an autocrine loop that also involved the RAS/RAF/MEK/ERK pathway (Lefevre et al, 2004). An incomplete response to c-Kit inhibition indicates that additional factors are involved (Hofmann et al, 2009). In addition, the GNAQ gene was shown to be mutated in almost half of UM (Van Raamsdonk et al, 2008). GNAQ is part of the G-protein heterotrimer and represents the GTP-binding part that couples GPCR signalling to MAPK activation, which marks it as a potential therapeutic target. However, targeting downstream signalling molecules may be just as effective as they may be shared with other mutant pathways. Tyrosine kinase activity profiling in UM was used to explore the involved kinases. On the basis of a UM cell line and two related metastasis cell lines, which revealed reduced ERK1/2 activation in metastases, we were able to identify Src as a crucial upstream tyrosine kinase for ERK1/2 activation in primary UM. Unfortunately, metastasis cell lines seemed less dependent on Src and may indicate that metastasis may require an alternative approach for intervention.
Materials and methods
Cell lines and tumour material
A total of 11 cell lines derived from primary UM (92.1; OCM-1, -3 and -8; Mel-202, -270, -285, -290) and UM metastases (OMM-1, -2.3 and -2.5) were analysed for kinase activity (Kan-Mitchell et al, 1989; Waard-Siebinga et al, 1995; Luyten et al, 1996; Chen et al, 1997). UM cell lines were cultured in RPMI 1640 medium (Gibco, Paisley, Scotland) supplemented with 3 mM L-glutamine (Gibco), 2% penicillin/streptomycin and 10% FBS (Hyclone, Logan, UT, USA). Primary UM was cultured in Amniochrome Pro Medium (Lonza Group Ltd, Basel, Switzerland). All cell cultures were incubated at 37°C in a humidified 5% CO2 atmosphere.
Cell lysates were obtained by lysing cells in M-PER Mammalian Protein Extract Reagent (Pierce, Rockford, IL, USA), supplemented with 1% Halt Protease Inhibitor Cocktail, EDTA-free (Pierce) and 1% Halt Phosphatase Inhibitor Cocktail (Pierce). Protein concentrations were measured by using the BCA Protein Assay kit (Pierce). Cell lysates were also acquired from three fresh primary UM samples obtained by enucleation and from three liver metastases of three different patients, in whom the diagnosis was confirmed.
Phospho-MAPK array
The Human Phospho-MAPK Array (R&D Systems, Abingdon, UK) was used to simultaneously detect the relative levels of nine MAP kinases and nine other serine/threonine kinases in cell lines, in primary UM and in liver metastasis. In this array, capture and control antibodies were spotted in duplicate on nitrocellulose membranes. Experiments were carried out according to the manufacturer's guidelines. In short, cell lysates were diluted and incubated with the array. After binding of both phosphorylated and unphosphorylated kinases, unbound material was washed away. A cocktail of phospho-site-specific biotinylated antibodies was used to detect phosphorylated proteins through streptavidin-horseradish peroxidase and chemiluminescence. X-ray films of the blots were scanned and analysed using G-boxHR (Syngene, Frederick, MD, USA). Control spots with mouse, goat and rabbit antibodies were used for background correction.
PamGene tyrosine kinase array
Experiments were carried out using a 4-array semi-automated system (PamStation 4, PamGene, 's-Hertogenbosch, The Netherlands) designed for processing PamChip-4 arrays. The PamChip Tyrosine Kinase Array (PamGene) contains 144 phospho-peptides, immobilised on a porous microarray surface through the peptide N terminus, representing tyrosine kinase substrates. Each array was blocked with 0.2% bovine serum albumin (BSA), fraction V (Calbiochem Immunochemicals, Merck KGaA, Darmstadt, Germany) by pumping it through the porous microarray for 30 cycles of 30 s. Thereafter, each array was washed thrice for 8 s with 1 × ABL Protein Tyrosine Kinase Reaction Buffer solution (New England Biolabs, Ipswich, MA, USA). Next, incubation was carried out at 30°C with the reaction mix, containing 5 μg cell lysate, 4 μl 100 × BSA (New England Biolabs), 0.4 μl 10 mM ATP (Sigma-Aldrich, Zwijndrecht, The Netherlands) and 0.5 μl 1 mg ml−1 monoclonal anti-phosphotyrosine FITC conjugate (clone Py20, Exalpha Biologicals, Maynard, MA, USA), adjusted to 40 μl with distilled H2O. The sample was pulsed back and forth through the porous material for 45 cycles, which is coupled to the base of a well to maximise reaction kinetics and to reduce analysis time. At every fifth pump cycle, a 16-bit TIFF image was taken with a built-in CCD camera (PamGene).
Blocking experiments were carried out with Src family-selective tyrosine kinase inhibitors, PP1, PP2 (Biomol International, LP, of Plymouth Meeting, PA, USA) and PP3 (the inactive analogue, Calbiochem), at an end concentration of 10 μ M in line with a large body of literature. Each particular inhibitor was mixed with lysates of cell lines and tissue, together with the reaction mix just before incubation on the array.
Acquired data from the PamStation 4 were captured with the supplied software package BioNavigator (Version 0.3.1; PamGene). For the purpose of finding differentially phosphorylated substrates, data were imported in the LIMMA package (Bioconductor.org) and we applied the empirical Bayes method (Smyth, 2004). Background subtracted data were normalised for differences between experiments, and substrates and P-values of 0.05 or less were corrected for multiple testing using the Benjamini and Hochberg correction. Substrates with a corrected P-value of 0.05 or less were assumed to be significant.
Western blot analysis
Cell lysates (10 μg) were separated on 12.5% SDS–PAGE gels, and proteins were transferred to Hybond-polyvinyldifluoride membranes (Amersham Biosciences, Buckinghamshire, UK). The membranes were blocked with 5% skim milk in a PBS-Tween 0.1% solution and probed at room temperature for 1 h with antibodies specific to each antigen: phospho-Src (Tyr527; dilution 1 : 1000), phospho-Src family (Tyr416; dilution 1 : 1000) and Src (36D10; dilution 1 : 1000) antibody (all from Cell Signaling Technology, Hertfordshire, UK). An antibody against actin (Abcam, Cambridge, UK) was used as a loading control. Membranes were subsequently incubated at room temperature with horseradish peroxidase-conjugated IgG anti-mouse or anti-rabbit secondary antibodies for 1 h. Supersignal West Femto ECL (Pierce) was used to visualise protein bands on the membrane.
siRNA treatment
Sub-confluent cell cultures were grown without antibiotics 24 h before transfection in RPMI 1640 medium. A mixture of Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) and two different siRNA constructs (40 nM) were incubated in the standard medium with reduced serum (1%), according to the manufacturer's instructions. The siRNA constructs (Stealth) were predesigned and validated (∼70% knockdown) by the manufacturer (Invitrogen). After 24 and 48 h, the cells were harvested and RNA and protein lysates were prepared.
WST-1 assay
Cell proliferation in response to PP1 (10 and 50 μ M) was measured by mitochondrial function using the WST-1 proliferation reagent (Sigma-Aldrich) as described earlier (Narayanan et al, 2005). This assay measures tetrazolium reductase activity in the mitochondria, which serves as a measure of cell viability. In short, 96-well plates were filled with 1250 UM cells per well. At 1 or 3 days (tumour 1–5) and 1 or 6 days (92.1; OCM-1, -3 and -8; Mel-202, -270, -285, -290, OMM-1, -2.3 and -2.5) after treatment, the WST-1 reagent was added and absorbance was measured at 450 nm on a multiwell spectrophotometer. The median and standard error of eight wells were taken at each time and dosage point.
Quantitative PCR
The cell lines (92.1; OCM-1, -3 and -8; Mel-202, -270, -285, -290, OMM-1, -2.3 and -2.5) were analysed for Src gene expression. Primers for Src and the reference gene, β-actin, were developed with Beacon Designer software (Premier Biosoft, Palo Alto, CA, USA). Primer sequences for Src: 5′-GCTGCGGCTGGAGGTCAAG-3′ (forward) and 5′-AGACATCGTGCCAGGCTTCAG-3′ (reverse). Primer sequence for β-actin: 5′-CGGGACCTGACTGACTACCTC-3′ (forward) and 5′-CTCCTTAATGTCACGCACGATTTC-3′ (reverse). The PCR reaction settings were 95°C for 5 min, then 40 cycles at 96°C for 15 s and at 60°C for 45 s. The DNA melting point of the amplicons was acquired by measuring the fluorescence of SYBR Green (Bio-Rad, Hercules, CA, USA) during a linear temperature transition from 70 to 97°C at 0.2°C each for 10 s with accompanying software (Bio-Rad).
Results
ERK1/2 activation in UM
An antibody array was applied to investigate the MAPK pathway in 10 UM cell lines, in three primary UM and three UM metastasis. We observed a uniform HSP27 phosphorylation, with the exception of in three UM cell lines (OCM1, -3, -8). UMs displaying activated ERK1/2 as well as phosphorylated HSP27 were most common, whereas signals for phosphorylated ERK1/2 were low in metastasis tissue (MET1-3) and metastatic UM cell lines (OMM1, OMM2.3 and OMM2.5) (Figure 1). Remarkably, two of the metastatic cell lines (OMM2.3, OMM2.5) are derived from the same patient as cell line Mel270 but contained far less activated ERK1/2.
The MAPK activation in primary UM and UM metastases was studied with a MAPK antibody array. We observed uniform HSP27 phosphorylation in both cell lines and tissue samples, except in OCM1, -3 and -8 (A). Activated ERK1/2 was normalised with HSP27 and shown to be low in UM metastases (MET1-3), whereas metastatic cell lines just passed the background (OMM1, -2.3, -2.5) (B).
Differential kinase activity in UM
Reduction of ERK1/2 activation in metastatic cell lines compared with that in primary UM cell lines provides a model to identify the underlying mechanism of ERK1/2 activation in the absence of BRAF and NRAS mutations.
To investigate whether a kinase is differentially activated between primary UM cell lines and metastatic UM cell lines, we used peptide-based tyrosine kinase arrays (Lemeer et al, 2007). The UM cell lines displayed a high kinase activity, whereas the metastatic UM cell lines displayed a low kinase activity, although the same amount of lysate was incubated (Figure 2A). After normalisation, we could analyse the kinase data and identify nine substrates that were significantly differentially phosphorylated between primary and metastatic UM cell lines (Figure 2B, Table 1). Primary UM and metastatic tissue also showed differential phosphorylation of these nine peptides, although not as clearly as observed in the cell lines (Figure 2C).
Tyrosine kinase activity was measured with an array of peptide substrates. Two representative examples of a UM cell line and a metastatic cell line (A). Analysis with eBayes identified nine substrates, representing eight proteins, to be significantly (P=0.01) differentially phosphorylated between UM and metastatic cell lines (B). UM (UM1-3) tyrosine kinase activity is high compared with liver metastasis (MET1-3) (two representative arrays are shown) (C).
Candidate kinase: Src
We identified nine peptides derived from eight proteins that were differentially phosphorylated between primary and metastatic cell line lysates. On the basis of a literature search, we identified candidate tyrosine kinases for eight out of nine peptides (Table 1) (Cooper et al, 1983; Thomas and Brugge, 1997; Koike et al, 2003; Diella et al, 2004). Among the candidates, Src and Src family members were most prominent. To validate the candidacy of Src, we performed in vitro inhibition experiments with the Src-kinase-specific inhibitors PP1 and the PP1 analogue, PP2. We added PP1 and PP2 (10 μ M) to lysates of primary UM tissue and of a primary UM cell line and measured the inhibitory effect of these Src inhibitors using kinase array (Figure 3A). A total of seven out of nine substrates that identified Src in the first screen displayed a significantly reduced phosphorylation when PP1 or PP2 were added to lysates of UM1 and Mel270 (Figure 3A). The PLCG1 peptide and one of the PAX1 (Y31) substrates did not reach significance but were still phosphorylated at a reduced level after PP1 and PP2 treatments. The peptide representing FAK1 Y576/Y577 is a genuine substrate for Src, which was not detected in the UM cell line comparison, but phosphorylation was significantly downregulated by PP1 and PP2 treatments. In the control experiment, in which we added the inactive analogue of PP1 (PP3) to cell lysates, we did not observe a loss of kinase activity (not shown).
UM1 (06-12) and Mel270 treatment with Src inhibitors (PP1/PP2) identified eight substrates with a significant reduction in phosphorylation (A). The inhibition of EFS peptide phosphorylation by genetic (Src siRNA) and pharmacological means (PP1) in cell line Mel270, and PP1 treatment of cell lysates of UM (UM1) and metastasis tissue (MET3) (B).
The kinase activity of metastasis tissue and UM tissue differed marginally (Figure 2C), and incubation with PP1 (10 μ M) resulted in a decimation of kinase activity similar to the inhibition that we observed in UM tissue (Figure 3B). To validate Src activity, Mel270 was transfected with two siRNA constructs that target Src and reduced kinase activity (Figure 3B).
Regulation of ERK1/2 and growth
To investigate whether Src contributes to ERK1/2 activation in Mel270, we analysed the two Src siRNA-transfected cell cultures with the MAPK antibody array. At 24 and 48 h after transfection with Src siRNA, we observed a reduced ERK1 phosphorylation, whereas ERK2 phosphorylation was minimally affected (Figure 4).
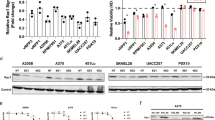
Whether Src inhibition and consequently a lowered ERK activation in UM cell lines is associated with a reduced growth was investigated with the WST-1 viability assay (Figure 5). All UM cell lines showed a PP1-dose and time (1–6 days)-dependent reduction in cell viability but the magnitude of the response differed widely. In general, the metastatic UM cell lines were less affected by PP1. We also determined the growth inhibition rate of PP1 in cultures of five primary UM cell cultures and observed an increased sensitivity to PP1 treatment compared with the cell lines. We had to take samples at day 3 of PP1 treatment because, thereafter, massive cell death occurred (Figure 5B).
UM cell lines and primary cultures were cultured with PP1 (10 μ M and 50 μ M). After 24 h and at 3 days (UM cultures) and 6 days (cell lines), viability was tested with the WST-1 assay. Two representative cell culture experiments for which all time points and conditions are shown (A). Growth inhibition by PP1 (50 μ M) after 24 h and at 3 and 6 days was normalised to the control culture of each individual cell line (B).
Src protein is reduced in metastasis cell line
Src is regulated by the phosphorylation of tyrosine residues at position 416 (Y416) and 527 (Y527). The expression of phosphorylated Src Y416, which is associated with an active conformation, was low in the metastatic UM cell lines (Figure 6A). Surprisingly, the phosphorylation of Y527, which is associated with an inactive conformation, was also low, and a subsequent analysis indicated that Src expression is low in metastatic UM cell lines. Therefore, the difference between kinase activity in metastatic cell lines (OMM1, OMM2.3 and OMM2.5) and UM cell lines (OCM1, OCM3, OCM8, Mel202, Mel270, Mel285, Mel290 and 92.1) seems to be the result of a difference in Src expression.
Western analysis of Src in the UM cell lines for activating phosphorylation (Y416), inactivating phosphorylation (Y527) and total Src expression (A). Src gene expression measured by qPCR varied widely but did not correlate with a variation in protein expression (B). UM and metastasis tissue all displayed a medium Src kinase expression, except for UM3, which presents a high level of expression (C).
To investigate the origin of a lowered Src expression, we performed a gene expression analysis (Figure 6B). Src gene expression varied widely in the cell lines and in the metastatic cell lines, but a correlation between protein and gene expression was not observed in UM cell lines.
A western analysis of Src expression in UM and metastasis tissue revealed a very high Src expression in only one out of three primary UM, whereas all three metastasis tissues displayed medium expression of Src protein (Figure 6C).
Discussion
Constitutive activation of ERK1/2 has often been reported for UM (Rimoldi et al, 2003; Weber et al, 2003; Zuidervaart et al, 2005). Using a more quantitative approach, we distinguished a decrease in active ERK1/2 in metastatic cell lines and in fresh liver metastasis, suggesting a loss of ERK1/2 activation during UM progression. The latter is unexpected as ERK1/2 activation is generally associated with mitogen signalling and is known to determine malignant potential in vitro. However, in endometrial and breast cancer, ERK1/2 activation has been associated with a good prognosis (Milde-Langosch et al, 2005) (Mizumoto et al, 2007). A possible explanation is provided by the observation that ERK1/2 is involved in oncogene and stress-induced senescence (Serrano et al, 1997; Stott et al, 1998). This mechanism is thought to be an important defence for cells that are at risk of neoplastic transformation and need to be circumvented by tumour cells in order to proliferate. Loss of activated ERK1/2 may not only relieve the associated inhibitory mechanisms in a direct manner but may also require alternative mitogenic signals to take over in UM metastasis.
Metastatic and UM cell lines provide a unique model to identify the mechanisms that regulate ERK1/2 activation in UM. Earlier work already showed that ERK1/2 phosphorylation in UM depends on the MAPK pathway, although mutations in the usual suspects (e.g., BRAF and NRAS) are lacking (Calipel et al, 2006). We investigated the possibility of a tyrosine kinase with differential activities in UM and UM metastasis to be responsible for ERK1/2 activation, using an array of kinase activity assays. Src was revealed as a differentially activated tyrosine kinase and this was supported by incubation with Src-specific kinase inhibitors, PP1 and PP2. Moreover, by treating cell lysates instead of cell cultures, we minimised the secondary effects of the inhibitors. However, PP1 and PP2 affect most of the Src family of tyrosine kinases, and the observed reduction in kinase activity therefore does not specifically mark Src. Multi-target inhibitors are a problem in molecular analysis but may be beneficial in clinical application, as, in CM, a switch from Src to Yes signalling has been reported in brain metastases (Summy and Gallick, 2003). To specifically inhibit Src, we targeted the Src gene expression with a siRNA approach. We detected a reduced kinase activity in Mel270 on transfection and this was correlated with a reduced ERK1 activation. The ERK2 activation seemed unaffected, which could be because of the limited efficacy of siRNA treatment, or it could indicate the activity of another, yet unidentified, kinase. A low Src protein expression in conjunction with a loss of ERK1/2 activation in metastatic UM cell lines, however, supports the hypothesis that, in UM, Src kinase is involved in ERK1/2 activation. Gene expression analysis revealed no significant differences between metastatic and UM cell lines, and thereby indicated that post-transcriptional mechanisms are most likely involved in Src downregulation. Src is both a kinase as well as a client protein for the chaperone HSP90 that is expressed in UM (Missotten et al, 2003). Whether HSP90 is reduced in metastases and whether treatment with HSP90 inhibitors depends on Src signalling is part of future investigation (Babchia et al, 2008; Faingold et al, 2008). The inhibition of Src kinase activity resulted in a strong growth reduction in all UM cultures, whereas in UM cell lines, the response varied more widely. The genetic background of the cell lines might play a role in the observed variation. However, all UM cell lines displayed Src kinase activity and PP1 sensitivity, irrespective of c-kit upregulation (Mel270) or the BRAF V600E (OCM1) and GNAQ Q209L (Mel202) mutation status (Lefevre et al, 2004; Van Raamsdonk et al, 2008). Tissue of UM and UM liver metastasis displayed more or less comparable Src levels. The incubation of lysates with Src inhibitors resulted in a comparable reduction of kinase activity in UM and metastasis tissue. The possibility that there exist Src negative clones in liver metastasis can, however, not be ruled out on the basis of these data. Clinical trials targeting Src kinase activity in UM should therefore anticipate this potential risk.
In conclusion, we have identified a differential ERK1/2 activation in UM and metastatic UM cell lines. Using tyrosine kinase activity profiling, we identified Src as a determinant of ERK1/2 activation and showed that Src expression and kinase activity, together with ERK1/2 activation, are reduced in UM metastases cell lines.
Change history
16 November 2011
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Babchia N, Calipel A, Mouriaux F, Faussat AM, Mascarelli F (2008) HSP90 inhibitors 17-AAG and 17-DMAG target wild-type B-Raf signaling for the proliferation of human uveal melanoma cell lines. Invest Ophthalmol Vis Sci 49: 2348–2356
Calipel A, Mouriaux F, Glotin AL, Malecaze F, Faussat AM, Mascarelli F (2006) Extracellular signal-regulated kinase-dependent proliferation is mediated through the protein kinase A/B-Raf pathway in human uveal melanoma cells. J Biol Chem 281: 9238–9250
Chen PW, Murray TG, Salgaller ML, Ksander BR (1997) Expression of MAGE genes in ocular melanoma cell lines. J Immunother 20: 265–275
Cohen Y, Goldenberg-Cohen N, Parrella P, Chowers I, Merbs SL, Pe′er J, Sidransky D (2003) Lack of BRAF mutation in primary uveal melanoma. Invest Ophthalmol Vis Sci 44: 2876–2878
Cooper JA, Reiss NA, Schwartz RJ, Hunter T (1983) Three glycolytic enzymes are phosphorylated at tyrosine in cells transformed by Rous sarcoma virus. Nature 302: 218–223
Cruz III F, Rubin BP, Wilson D, Town A, Schroeder A, Haley A, Bainbridge T, Heinrich MC, Corless CL (2003) Absence of BRAF and NRAS mutations in uveal melanoma. Cancer Res 63: 5761–5766
Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E, Ewing R, Floyd Y, Gray K, Hall S, Hawes R, Hughes J, Kosmidou V, Menzies A, Mould C, Parker A, Stevens C, Watt S, Hooper S, Wilson R, Jayatilake H, Gusterson BA, Cooper C, Shipley J, Hargrave D, Pritchard-Jones K, Maitland N, Chenevix-Trench G, Riggins GJ, Bigner DD, Palmieri G, Cossu A, Flanagan A, Nicholson A, Ho JW, Leung SY, Yuen ST, Weber BL, Seigler HF, Darrow TL, Paterson H, Marais R, Marshall CJ, Wooster R, Stratton MR, Futreal PA (2002) Mutations of the BRAF gene in human cancer. Nature 417: 949–954
Diella F, Cameron S, Gemund C, Linding R, Via A, Kuster B, Sicheritz-Ponten T, Blom N, Gibson TJ (2004) Phospho.ELM: a database of experimentally verified phosphorylation sites in eukaryotic proteins. BMC Bioinformatics 5: 79
Edmunds SC, Cree IA, Di Nicolantonio F, Hungerford JL, Hurren JS, Kelsell DP (2003) Absence of BRAF gene mutations in uveal melanomas in contrast to cutaneous melanomas. Br J Cancer 88: 1403–1405
Egan KM, Seddon JM, Glynn RJ, Gragoudas ES, Albert DM (1988) Epidemiologic aspects of uveal melanoma. Surv Ophthalmol 32: 239–251
Faingold D, Marshall JC, Antecka E, Di Cesare S, Odashiro AN, Bakalian S, Fernandes BF, Burnier Jr MN (2008) Immune expression and inhibition of heat shock protein 90 in uveal melanoma. Clin Cancer Res 14: 847–855
Goding CR (2000) Mitf from neural crest to melanoma: signal transduction and transcription in the melanocyte lineage. Genes Dev 14: 1712–1728
Hofmann UB, Kauczok-Vetter CS, Houben R, Becker JC (2009) Overexpression of the KIT/SCF in uveal melanoma does not translate into clinical efficacy of imatinib mesylate. Clin Cancer Res 15: 324–329
Janssen CS, Sibbett R, Henriquez FL, McKay IC, Kemp EG, Roberts F (2008) The T1799A point mutation is present in posterior uveal melanoma. Br J Cancer 99: 1673–1677
Kan-Mitchell J, Mitchell MS, Rao N, Liggett PE (1989) Characterization of uveal melanoma cell lines that grow as xenografts in rabbit eyes. Invest Ophthalmol Vis Sci 30: 829–834
Koike A, Kobayashi Y, Takagi T (2003) Kinase pathway database: an integrated protein-kinase and NLP-based protein-interaction resource. Genome Res 13: 1231–1243
Lefevre G, Glotin AL, Calipel A, Mouriaux F, Tran T, Kherrouche Z, Maurage CA, Auclair C, Mascarelli F (2004) Roles of stem cell factor/c-Kit and effects of Glivec/STI571 in human uveal melanoma cell tumorigenesis. J Biol Chem 279: 31769–31779
Lemeer S, Jopling C, Naji F, Ruijtenbeek R, Slijper M, Heck AJ, den Hertog J (2007) Protein-tyrosine kinase activity profiling in knock down zebrafish embryos. PLoS ONE 2: e581
Luyten GP, Naus NC, Mooy CM, Hagemeijer A, Kan-Mitchell J, Van Drunen E, Vuzevski V, de Jong PT, Luider TM (1996) Establishment and characterization of primary and metastatic uveal melanoma cell lines. Int J Cancer 66: 380–387
Maat W, Kilic E, Luyten GP, de Klein A, Jager MJ, Gruis NA, van der Velden PA (2008) Pyrophosphorolysis detects B-RAF mutations in primary uveal melanoma. Invest Ophthalmol Vis Sci 49: 23–27
Milde-Langosch K, Bamberger AM, Rieck G, Grund D, Hemminger G, Muller V, Loning T (2005) Expression and prognostic relevance of activated extracellular-regulated kinases (ERK1/2) in breast cancer. Br J Cancer 92: 2206–2215
Missotten GS, Journee-de Korver JG, Wolff-Rouendaal D, Keunen JE, Schlingemann RO, Jager MJ (2003) Heat shock protein expression in the eye and in uveal melanoma. Invest Ophthalmol Vis Sci 44: 3059–3065
Mizumoto Y, Kyo S, Mori N, Sakaguchi J, Ohno S, Maida Y, Hashimoto M, Takakura M, Inoue M (2007) Activation of ERK1/2 occurs independently of KRAS or BRAF status in endometrial cancer and is associated with favorable prognosis. Cancer Sci 98: 652–658
Mooy CM, Van der Helm MJ, Van der Kwast TH, de Jong PT, Ruiter DJ, Zwarthoff EC (1991) No N-ras mutations in human uveal melanoma: the role of ultraviolet light revisited. Br J Cancer 64: 411–413
Narayanan R, Kenney MC, Kamjoo S, Trinh TH, Seigel GM, Resende GP, Kuppermann BD (2005) Trypan blue: effect on retinal pigment epithelial and neurosensory retinal cells. Invest Ophthalmol Vis Sci 46: 304–309
Reddy KB, Nabha SM, Atanaskova N (2003) Role of MAP kinase in tumor progression and invasion. Cancer Metastasis Rev 22: 395–403
Rimoldi D, Salvi S, Lienard D, Lejeune FJ, Speiser D, Zografos L, Cerottini JC (2003) Lack of BRAF mutations in uveal melanoma. Cancer Res 63: 5712–5715
Satyamoorthy K, Li G, Gerrero MR, Brose MS, Volpe P, Weber BL, Van Belle P, Elder DE, Herlyn M (2003) Constitutive mitogen-activated protein kinase activation in melanoma is mediated by both BRAF mutations and autocrine growth factor stimulation. Cancer Res 63: 756–759
Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW (1997) Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88: 593–602
Singh AD, Topham A (2003) Incidence of uveal melanoma in the United States: 1973–1997. Ophthalmology 110: 956–961
Smyth GK (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3, (Article 3)
Soparker CN, O’Brien JM, Albert DM (1993) Investigation of the role of the ras protooncogene point mutation in human uveal melanomas. Invest Ophthalmol Vis Sci 34: 2203–2209
Stott FJ, Bates S, James MC, McConnell BB, Starborg M, Brookes S, Palmero I, Ryan K, Hara E, Vousden KH, Peters G (1998) The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J 17: 5001–5014
Summy JM, Gallick GE (2003) Src family kinases in tumor progression and metastasis. Cancer Metastasis Rev 22: 337–358
Thomas SM, Brugge JS (1997) Cellular functions regulated by Src family kinases. Annu Rev Cell Dev Biol 13: 513–609
van der Velden PA, Metzelaar-Blok JA, Bergman W, Monique H, Hurks H, Frants RR, Gruis NA, Jager MJ (2001) Promoter hypermethylation: a common cause of reduced p16(INK4a) expression in uveal melanoma. Cancer Res 61: 5303–5306
van Elsas A, Zerp S, van der FS, Kruse-Wolters M, Vacca A, Ruiter DJ, Schrier P (1995) Analysis of N-ras mutations in human cutaneous melanoma: tumor heterogeneity detected by polymerase chain reaction/single-stranded conformation polymorphism analysis. Recent Results Cancer Res 139: 57–67
Van Raamsdonk CD, Bezrookove V, Green G, Bauer J, Gaugler L, O’Brien JM, Simpson EM, Barsh GS, Bastian BC (2008) Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature 457 (7229): 599–602
Waard-Siebinga I, Blom DJ, Griffioen M, Schrier PI, Hoogendoorn E, Beverstock G, Danen EH, Jager MJ (1995) Establishment and characterization of an uveal-melanoma cell line. Int J Cancer 62: 155–161
Weber A, Hengge UR, Urbanik D, Markwart A, Mirmohammadsaegh A, Reichel MB, Wittekind C, Wiedemann P, Tannapfel A (2003) Absence of mutations of the BRAF gene and constitutive activation of extracellular-regulated kinase in malignant melanomas of the uvea. Lab Invest 83: 1771–1776
Zuidervaart W, van Nieuwpoort F, Stark M, Dijkman R, Packer L, Borgstein AM, Pavey S, van der Velden PA, Out C, Jager MJ, Hayward NK, Gruis NA (2005) Activation of the MAPK pathway is a common event in uveal melanomas although it rarely occurs through mutation of BRAF or RAS. Br J Cancer 92: 2032–2038
Acknowledgements
We acknowledge the technical assistance of Mieke Versluis, Amal F Abukar, Aabed Baghat and the staff at PamGene. We also thank Jolanda Reek for statistical analysis. This study was supported by Grant RUL 2001-2472 from the Dutch Cancer Society (KWF).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Maat, W., Filali, M., Mulder, A. et al. Episodic Src activation in uveal melanoma revealed by kinase activity profiling. Br J Cancer 101, 312–319 (2009). https://doi.org/10.1038/sj.bjc.6605172
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.bjc.6605172