Abstract
Maspin, a member of the serpin family, has a role in cell migration, angiogenesis and apoptosis. Little is known of the clinical significance of maspin gene expression in human cancers. We developed a real-time quantitative RT–PCR assay to quantify the full range of maspin mRNA copy numbers in a series of 10 ERα-positive and 10 ERα-negative breast tumours. We observed a statistical link between low maspin mRNA levels and positive oestrogen status (P=0.0012). In consequence, to better assess the prognostic value of maspin gene expression in breast cancer, we then quantified maspin mRNA content in an additional independent well-defined cohort of 105 ERα-positive postmenopausal breast cancer patients treated with primary surgery followed by adjuvant tamoxifen alone. Maspin expression varied widely in tumour tissues (by nearly four orders of magnitude), being underexpressed in 33 out of 105 tumours (31.4%) and overexpressed in 24 out of 105 tumours (22.9%) relative to normal breast tissues. Immunohistochemical studies demonstrated that maspin protein was strictly expressed in myoepithelial cells of normal breast tissue and in tumour epithelial cells, exclusively in maspin-overexpressing tumours. Patients with tumours overexpressing the maspin gene had significantly shorter relapse-free survival after surgery than patients whose tumours normally expressed or underexpressed maspin (P=0.0011). The prognostic significance of maspin overexpression persisted in Cox multivariate regression analysis (P=0.0024). These findings show that the maspin mRNA level can have important prognostic significance in human breast cancer, and point to the maspin gene as a putative molecular predictor of hormone responsiveness in breast cancer.
Similar content being viewed by others
Main
Maspin (mammary serpin) is a 42-kDa cytoplasmic protein belonging to the serpin family of serine protease inhibitors (Zou et al, 1994). The maspin gene was originally identified in normal mammary epithelium by subtractive hybridisation on the basis of its expression at the mRNA level (Zou et al, 1994). Maspin has subsequently been localised to epithelial and myoepithelial cells in a variety of tissues (Pemberton et al, 1997).
Biological studies suggest a tumour-suppressive role of maspin, mediated by effects on cell migration (Zou et al, 1994; Sheng et al, 1996), angiogenesis (Zhang et al, 2000a) and apoptosis (Zhang et al, 2000b; Jiang et al, 2002). Maspin expression in breast tumour cells reduces tumour induction and metastasis in nude mice (Zou et al, 1994; Shi et al, 2001; Streuli, 2002), and also invasion of the basement membrane in vitro (Zou et al, 1994). Moreover, treatment of human breast cancer cells with recombinant maspin (rMaspin) inhibits cell motility (Sheng et al, 1996). Possible molecular targets for maspin are tissue-type plasminogen activator (tPA) (Sheng et al, 1998) and various integrins (Seftor et al, 1998).
The role of maspin in human breast cancer is poorly documented, and even less is known about the clinical significance of maspin expression in this setting. The few available data on maspin expression in breast cancer have been obtained using breast cancer cell lines and breast tumour series (Zou et al, 1994; Jiang et al, 1997; Domann et al, 2000; Maass et al, 2001a), and the results are confusing. Indeed, maspin expression is generally downregulated in primary breast tumours and lost in mammary carcinoma lines (Zou et al, 1994; Hojo et al, 2001; Maass et al, 2001a, 2001b), but some infiltrating breast tumours have been found to show strong upregulation (Hojo et al, 2001; Maass et al, 2001b). Maspin expression has been found to decrease with increasing malignancy in primary breast tumours, and to be absent from distant metastases (Zou et al, 1994). Surprisingly, Martin et al (2000), using cDNA array technology, identified the maspin gene as the leader of a cluster of genes that are strongly upregulated in ERα-negative breast tumours (ERα negativity is associated with aggressive tumours). Finally, four recent clinical studies of heterogeneous series of women with breast tumours examined the possible association between maspin expression (mRNA or protein level) and classical clinical and pathological parameters, including patient outcome (Hojo et al, 2001; Maass et al, 2001a, 2001b). The results of these studies are conflicting, three studies suggested that maspin underexpression could be a potential poor prognostic marker in breast cancer, while the fourth identified maspin overexpression rather as an independent poor prognostic indicator in breast cancer patients (Umerika et al, 2002).
Here, we developed a real-time quantitative RT–PCR assay to quantify maspin mRNA in homogeneous total RNA solutions obtained from human tissue samples. As we found that maspin gene expression was strongly linked to ERα expression status, we quantified maspin mRNA expression in a well-defined cohort of 105 ERα-positive postmenopausal breast cancer patients who were treated with primary surgery, followed by adjuvant tamoxifen alone and whose long-term outcome was known. The significance of maspin as a prognostic factor is discussed.
Materials and methods
Patients and samples
We analysed tissue samples from primary breast tumours excised from 105 women at Centre René Huguenin from 1980 to 1994. Tumour tissue samples of the 105 patients were collected in accordance with French regulations.
Immediately following surgery, the tumour samples were stored in liquid nitrogen until RNA extraction. The patients (mean age 70.8 years, range 54–86) met the following criteria: primary unilateral nonmetastatic postmenopausal breast carcinoma; oestrogen receptor alpha positivity (as determined at the protein level by biochemical methods (dextran-coated charcoal method until 1988 and thereafter EIA) and confirmed by ERα real-time quantitative RT–PCR assay); complete clinical, histological and biological information available; no radiotherapy or chemotherapy before surgery; and full follow-up at Centre René Huguenin. Among the 105 breast tumours, 97 were invasive ductal carcinomas and eight were invasive lobular carcinomas. The standard prognostic factors are presented in Table 1. A total of 34 patients (32.4%) had modified radical mastectomy and 71 (67.6%) had breast-conserving surgery plus locoregional radiotherapy. Patients underwent physical examinations and routine chest radiography every 3 months for 2 years, then annually. Mammograms were performed annually. The median follow-up was 6.0 years (range 1.5–10.0 years). All the patients received postoperative adjuvant endocrine therapy (tamoxifen, 20 mg daily for 3–5 years), and no other treatment. In all, 31 patients relapsed (the distribution of first relapse events was as follows: 27 metastases, and four both local and/or regional recurrences and metastases).
To investigate the relation between maspin mRNA levels and ERα expression status, we also analysed 20 additional primary breast tumours: 10 ERα-negative and 10 ERα-positive tumours.
We analysed six breast tumuor cell lines obtained from the American Tissue Type Culture Collection (SK-BR-3, T-47D, BT-20, HBL-100, ZR-75-1 and MCF7).
Specimens of adjacent normal breast tissue from five of the breast cancer patients, and normal breast tissue from three women undergoing cosmetic breast surgery, were used as sources of normal RNA.
Real-time RT–PCR
Theoretical basis
Reactions are characterised by the point during cycling when amplification of the PCR product is first detected, rather than the amount of PCR product accumulated after a fixed number of cycles. The higher the starting copy number of the target transcript, the earlier a significant increase in fluorescence is observed. The parameter Ct (threshold cycle) is defined as the fractional cycle number at which the fluorescence generated by the incorporation of a fluorogenic molecule in double-stranded DNA passes a fixed threshold above baseline.
The precise amount of total RNA added to each reaction mix (based on optical density) and its quality (i.e. lack of extensive degradation) are both difficult to assess. We therefore also quantified transcripts of the gene coding for the TATA box-binding protein (TBP) (a component of the DNA-binding protein complex TFIID) as the endogenous RNA control, and each sample was normalised on the basis of its TBP content.
The relative maspin gene expression level was also normalised to a calibrator, or 1 × sample, consisting of a pool of normal breast tissue specimens.
Final results, expressed as N-fold differences in maspin gene expression relative to the TBP gene and normal breast tissues (the calibrator), termed ‘Nmaspin’, were determined as follows:
where ΔCt values of the sample and calibrator are determined by subtracting the average Ct value of the maspin gene from the average Ct value of the maspin gene.
Primers and PCR consumables
Primers for the TBP and maspin genes were chosen with the assistance of the computer programs Oligo 4.0 (National Biosciences, Plymouth, MN, USA). We conducted BLASTN searches against dbEST, htgs and nr (the nonredundant set of the GenBank, EMBL and DDBJ database sequences) to confirm the total gene specificity of the nucleotide sequences chosen for the primers, and the absence of DNA polymorphisms. The nucleotide sequences of the primers used were as follows: Maspin-U (5′-CTA CTT TGT TGG CAA GTG GAT GAA-3′) and Maspin-L (5′-ACT GGT TTG GTG TCT GTC TTG TTG-3′) for maspin gene (PCR product of 90 bp), and TBP-U (5′-TGC ACA GGA GCC AAG AGT GAA-3′) and TBP-L (5′-CAC ATC ACA GCT CCC CAC CA-3′) for TBP gene (PCR product of 132 bp).
To avoid amplification of contaminating genomic DNA, one of the two primers was placed in a different exon. For example, the upper primer of TBP was placed at the junction between exons 5 and 6, whereas the lower primer was placed in exon 6. Agarose gel electrophoresis allowed us to verify the specificity of PCR amplicons.
RNA extraction
Total RNA was extracted from breast specimens by using the acid-phenol guanidium method. The quality of the RNA samples was determined by electrophoresis through agarose gels and staining with ethidium bromide, and the 18S and 28S RNA bands were visualised under ultraviolet light.
cDNA synthesis
RNA was reverse transcribed in a final volume of 20 μl containing 1 × RT buffer (500 mM each dNTP, 3 mM MgCl2, 75 mM KCl, 50 mM Tris-HCl pH 8.3), 10 units of RNasinTM Ribonuclease inhibitor (Promega, Madison, WI, USA), 10 mM dithiothreitol, 50 U of Superscript II RNase H-reverse transcriptase (Gibco BRL, Gaithersburg, MD, USA), 1.5 mM random hexamers (Pharmacia, Uppsala, Sweden) and 1 μg of total RNA. The samples were incubated at 20°C for 10 min and 42°C for 30 min, and reverse transcriptase was inactivated by heating at 99°C for 5 min and cooling at 5°C for 5 min.
PCR amplification
All PCR reactions were performed using an ABI Prism 7700 Sequence Detection System (Perkin-Elmer Applied Biosystems, Foster City, CA). Polymerase chain reaction was performed using the SYBR® Green PCR Core Reagents kit (Perkin-Elmer Applied Biosystems, Foster City, CA). The thermal cycling conditions comprised an initial denaturation step at 95°C for 10 min and 50 cycles at 95°C for 15 s and 65°C for 1 min. Experiments were performed with duplicates for each data point.
Immunohistochemical studies
Indirect immunoperoxidase staining of fixed tissues was performed using mouse monoclonal antibody G167-70 directed against the human maspin protein (PharMingen, San Diego, CA) and monoclonal antibody SMMS-1 raised against human myosin heavy chain (Dakocytomation Denmark A/S, Glostrup, DK).
The immunohistochemical procedure was applied to paraffin-embedded tissue sections. A water bath antigen-retrieval technique was used in all cases. Sections were mounted on precoated slides (Dako) and allowed to dry at 50°C overnight. The sections were then dewaxed in xylene and hydrated through graded dilutions of ethanol. Endogenous activity was blocked with 1% hydrogen peroxide for 15 min. Sections were then immersed in a heat-resistant plastic box containing 10 ml of pH 6.0 citrate buffer and processed in the water bath for 40 min. Sections were then allowed to cool to room temperature for 20 min before rinsing in H2O. The blocking reagent was poured off and the primary antibodies were left for 25 min. A standard avidin–biotin–peroxidase complex (LSAB) method was used to reveal the antibody–antigen reaction (Dako).
The localisation and intensity of staining were assessed by two independent pathologists who were blinded to the real-time RT–PCR results.
Statistical analysis
Relapse-free survival (RFS) was determined as the interval between diagnosis and detection of the first relapses (local and/or regional recurrences, and/or metastases).
Clinical, histological and biological parameters were compared using the χ2 test. Differences between the two populations were judged significant at confidence levels greater than 95% (P<0.05). The relation between maspin mRNA levels and ERα status was assessed using the Kruskal–Wallis test. Survival distributions were estimated by the Kaplan-Meier method (Kaplan and Meier, 1958), and the significance of differences between survival rates was ascertained using the log-rank test (Peto et al, 1977). Cox's proportional hazards regression model (Cox, 1972) was used to assess prognostic significance.
Results
Maspin mRNA expression in normal breast tissues
To determine the cutoff point for altered maspin expression in breast cancer tissue, the Nmaspin value, calculated as described in Patients and Samples, was determined for eight normal breast RNA samples (Table 2). As this value consistently fell between 0.59 and 1.67 (mean 1.04±s.d. 0.35), values of 3 (mean±5 s.d.) or more were considered to represent marked overexpression, and values of 0.30 (mean−2 s.d.) or less were considered to represent underexpression of maspin mRNA. We have previously used the same approach to determine the cutoff points for altered expression of other tumour genes (Bièche et al, 1999a).
Relation between maspin mRNA levels and ERα expression status
Maspin mRNA levels were determined in 10 ERα-negative and 10 selected ERα-positive breast tumours with very high ERα mRNA values (Table 2). These 10 ERα-negative and 10 ERα-positive breast tumours have been selected from a previously tested breast tumours series where the ERα status was determined at the protein level by biochemical methods and quantified by ERα real-time quantitative RT–PCR assay (Bièche et al, 2001).
We found a negative correlation between maspin and ERα mRNA expression (P=0.0012): the 10 ERα-negative breast tumours had higher levels of maspin mRNA (0.89- to 96.8-fold normal; mean 15.76±s.d. 30.87) than the 10 ERα-positive breast tumours (0.02- to 3.89-fold; mean 0.87±s.d. 1.12) (Table 2).
Maspin mRNA expression in a well-defined set of 105 ERα-positive postmenopausal breast tumour RNA samples
To better assess the prognostic value of maspin gene expression in ERα-positive breast cancer, we quantified maspin mRNA content in an additional independent well-defined cohort of 105 ERα-positive postmenopausal breast cancer patients treated with primary surgery followed by adjuvant tamoxifen alone. These breast tumour RNA samples had a wide range of Nmaspin values (0.01–46.7, i.e. nearly four orders of magnitude).
Compared to normal breast tissues, 57 (54.3%) tumours showed altered maspin mRNA expression. A total of 33 tumours (31.4%) showed maspin mRNA underexpression (Nmaspin from 0.01 to 0.28) and 24 (22.9%) showed maspin mRNA overexpression (Nmaspin from 3.11 to 46.7).
maspin expression was also investigated in six breast tumour cell lines. All showed maspin underexpression, with Nmaspin values of 0.004 (SK-BR-3), 0.029 (T-47D), 0.045 (BT-20), 0.001 (HBL-100), 0.042 (ZR-75-1) and 0.017 (MCF7).
Localisation of maspin protein in epithelial tumour cell cytoplasm
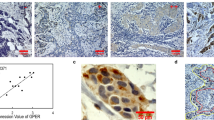
We detected specific maspin immunoreactivity in normal myoepithelial cells of all eight tumour samples studied by IHC. We also detected strong specific immunoreactivity in epithelial cells of the four tumours which overexpressed maspin mRNA and in none of the four tumours which did not overexpress maspin. We thus obtained a perfect match between maspin mRNA overexpression and IHC positivity (Figure 1). Maspin immunoreactivity was exclusively found in normal myoepithelial cells and tumour epithelial cells; infiltrating lymphocytes and normal glandular cells in the tumour were consistently negative. Staining was found exclusively in the cytoplasm of both normal myoepithelial and tumour epithelial cells (Figure 1). The positive tumour epithelial cells were myosin heavy-chain-negative (Figure 1), and were therefore not of myoepithelial origin.
Immunohistochemical staining for maspin (A, B, E, F) and for myosin heavy chain (C, D) in two breast tumours, one with normal maspin expression (A, C, E) and one with maspin overexpression (B, D, F). Intense maspin immunoreactivity was found in tumour epithelial cells from the maspin mRNA-overexpressing tumour (B) but not in cells (star) from the tumour without maspin overexpression (A) (original magnification × 20). Note the negative myosin heavy-chain staining of tumour epithelial cells from the maspin mRNA-overexpressing tumour (D), confirming that the latter is not of myoepithelial origin (original magnification × 20). In both cases, normal breast tissue showed maspin immunoreactivity in normal myoepithelial cells (arrows) (A, B), which also showed strong immunoreactivity for myosin heavy chain (C, D) (original magnification × 20). Maspin immunoreactivity was localised in the cytoplasmic compartment of both epithelial tumour cells (F) and normal myoepithelial cells (E) (original magnification × 100).
Correlation between maspin mRNA levels and clinical, pathological and biological parameters
We sought links between maspin mRNA expression status and standard clinical and pathological factors in breast cancer (Table 3). The only statistically significant association (χ2 test) was between maspin mRNA overexpression and Scarff–Bloom–Richardson (SBR) histopathological grade III (P=0.03). maspin mRNA status was not significantly associated with age, lymph node status, macroscopic tumour size or histological type.
We also found no link between maspin mRNA status and the mRNA status of two candidate genes predicting the response to endocrine therapy, namely ERBB2 and ERβ (estrogen receptorβ) (Table 3).
Prognosis analysis of maspin mRNA expression
Univariate analysis (log-rank test) showed that RFS was linked to maspin mRNA status (P=0.0011; Figure 2). The RFS of the 24 patients with maspin-overexpressing tumours (5-year RFS 41.8±10.5%) was shorter than that of the 48 patients whose tumours normally expressed maspin (5-year RFS 84.4±5.4%) and also shorter than that of the 33 patients with maspin-underexpressing tumours (5-year RFS 83.6±6.7%).
Interestingly, the outcome of the patients with maspin-overexpressing tumours was also significantly worse when the analysis focused on two well-known specific subsets of poor-prognosis breast cancer patients, that is, those with more than three involved lymph nodes (P=0.0049, Figure 3A) and those with histopathological grade III (P=0.035, Figure 3B).
Finally, the prognostic significance of the four parameters identified in univariate analysis (histopathological grade III, lymph-node status, macroscopic tumour size (Table 1) and maspin overexpression status (Table 1, Figure 2)) persisted (except for macroscopic tumour size) in Cox multivariate regression analysis of RFS (Table 4).
Discussion
Since 1994, when maspin was first described as a tumour-suppressor gene in breast cancer, little has been learnt about its expression. Maspin protein is present in the breast epithelium, and particularly in the myoepithelium, and maspin expression has been reported to be both decreased and increased in subsets of breast tumours, and lost in some metastases (Zou et al, 1994; Jiang et al, 1997; Domann et al, 2000). Four clinical studies of maspin expression have recently been reported, and these mainly focused on underexpression status, one at the mRNA level (Maass et al, 2001a) and three at the protein level (Hojo et al, 2001; Maass et al, 2001b; Umerika et al, 2002). Maass et al (2001a), using a semiquantitative nested RT–PCR assay in a small series of primary breast carcinoma tissues (n=45) with short follow-up (3 years), showed no maspin transcripts in 16 (36%) breast tumours. Maspin mRNA expression was unrelated to established prognostic factors, including ERα status, but six of the eight patients who developed distant metastasis within 3 years of the initial diagnosis showed no maspin mRNA expression in their primary breast tumours. Two immunochemical studies, although involving heterogeneous breast tumours series with no follow-up, also suggest that maspin underexpression could be a marker of breast cancer aggressiveness (Hojo et al, 2001; Maass et al, 2001b). In contrast, the third study identified maspin overexpression as an independent poor prognostic indicator in breast cancer patients (Umerika et al, 2002).
We used real-time quantitative RT–PCR to assess maspin gene expression in human breast tumours. This recent approach to nucleic acid quantification is suited to the development of target gene assays, having a high degree of inter-laboratory standardisation and yielding statistical confidence values (Bièche et al, 1999b). The main advantage of real-time RT–PCR is its large linear dynamic range. This made it particularly appropriate for analysing maspin gene expression, which ranged from 0.01 to 46.7 times normal in this series. Contrary to Maass et al (2001a), who failed to detect maspin mRNA in 36% of their breast cancer specimens with a semiquantitative nested RT–PCR assay, we detected maspin mRNA in all the breast tumour samples tested, probably reflecting the higher sensitivity of our real-time RT–PCR method.
It has been suggested that maspin dysregulation in tumours may be because of altered expressions of various transregulators acting on the maspin promoter, including the proteins Ets and Ap1 (Zhang et al, 1997) and p53 (Zou et al, 2000). A recent publication also discussed the involvement of aberrant cytosine methylation and chromatin condensation of the maspin promoter in the silencing of maspin expression during neoplastic progression (Domann et al, 2000).
We found a strong negative correlation between maspin gene expression and ERα expression status (Table 2), in agreement with Umerika et al (2002) and Martin et al (2000). These latter authors, using cDNA array technology, identified maspin gene as the leader of a cluster of genes that are strongly upregulated in ERα-negative breast tumours. Thus, to test the prognostic value of maspin mRNA expression in breast cancer, we chose to analyse a well-defined cohort of 105 postmenopausal patients with ERα-positive breast cancer treated with primary surgery followed by adjuvant tamoxifen alone.
We observed both underexpression (31.4% of samples) and overexpression (22.9%) of maspin mRNA.
In all, 33 tumours, and all six breast tumour cell lines tested (SK-BR-3, 47D, BT-20, HBL-100, ZR-75-1 and MCF7) underexpressed maspin mRNA. These results are in agreement with reports from several authors (Zou et al, 1994; Domann et al, 2000; Maass et al, 2001a) showing maspin underexpression at the RNA and/or protein level in breast cancer, and corroborating biological evidence that maspin is a tumour-suppressor gene. The very low maspin mRNA values in the six breast tumour cell lines tested by real-time RT–PCR were in total agreement with previous data (Zou et al, 1994; Zhang et al, 1997; Domann et al, 2000; Maass et al, 2001a). These very low values observed in the tumour cell lines could be because of the depletion in the in vitro epithelial breast tumour cell cultures of the myoepithelial normal cells, which secrete high levels of maspin (Sternlicht and Barsky, 1997). Contrary to Maass et al (2001a), but in agreement with Martin et al (2000) and Umerika et al (2002), we found an association between maspin mRNA underexpression and oestrogen receptor alpha positivity, suggesting that maspin mRNA underexpression is associated with low tumour aggressiveness (Table 2).
Finally, RFS after surgery was not shorter in patients whose tumours underexpressed maspin than in patients whose tumours normally expressed maspin (Figure 2). Zou et al (1994) and Sheng et al (1996) reported that maspin inhibited the invasive and metastatic potential of breast cancer cells, while Maass et al (2001a) found that maspin downregulation was associated with a higher risk of early distant metastasis in a series of breast tumour patients with short median follow-up (3 years).
We also identified 24 tumours with clear maspin overexpression, mRNA expression ranged from 3.1- to 47-fold that was found in normal breast tissue. Several authors have previously reported cases of maspin-overexpressing breast tumours (Hojo et al, 2001; Maass et al, 2001b; Umerika et al, 2002). By using immunohistochemical analysis, we showed that maspin transcripts are translated into maspin protein (Figure 1). We also showed a positive correlation between maspin mRNA overexpression and maspin protein abundance, and precised that alteration of maspin gene expression was exclusively found inside tumour epithelial cells (Figure 1). Taken together, these findings suggest that maspin gene expression is mainly dysregulated at the transcriptional level in breast cancer. It is also noteworthy that we found weaker maspin immunoreactivity in normal myoepithelial cells located in adjacent normal breast tissue found around all the tumour samples tested, in keeping with the literature (Sternlicht and Barsky, 1997).
In our ERα-positive postmenopausal breast tumour series, maspin mRNA overexpression was associated with poor long-term patient outcome relative to tumours with normal or low maspin expression (Figure 2). We also obtained evidence that maspin mRNA overexpression might be a further marker of poor outcome in patient subsets with classical markers of poor prognosis (more than three involved lymph nodes, or histopathological grade III) (Figure 3).
Primary surgery followed by adjuvant tamoxifen alone is the usual treatment for ERα-positive postmenopausal breast cancer. Our results, obtained in a well-defined cohort of ERα-positive postmenopausal breast cancer patients treated in this way, also suggest that maspin status, by evenly dichotomising such patients, might emerge as a useful predictor of the response to endocrine therapy. Moreover, the predictive power of maspin expression appears to be independent of two other genes, ERBB2 and ERβ, which have been proposed to predict the response to endocrine therapy (Speirs et al, 1999; Stal et al, 2000). Indeed, we found no link between maspin mRNA status and the mRNA status of these two genes (Table 3). Confirmation of the predictive value of maspin parameter in the response to endocrine therapy in breast cancer patients needs a prospective randomised study to show that this parameter do influence the outcome only in patients who received adjuvant tamoxifen as compared with untreated patients.
This relation between maspin gene overexpression and poor outcome in breast cancer is in agreement with Umerika et al (2002), but conflicts with the reported role of maspin as a tumour-suppressor gene that inhibits cell migration and angiogenesis. Maass et al (2001c), in a series of pancreatic tumours, found that maspin expression was gradually upregulated with increasing malignancy, from normal pancreas tissue to precancerous lesions and invasive carcinoma. One possible explanation for these paradoxical results is that maspin expression is upregulated in order to neutralise for overexpression of tumour-activating gene targets. It is noteworthy in this respect that other inhibitors of tumour progression, such as PAI-1 and PAI-2 (plasminogen activator inhibitor type 1 and type 2) are overexpressed in breast cancer and associated with a poor prognosis (Stephens et al, 1998).
These results confirm the interest to investigate maspin gene expression for the detection of circulating breast tumour cells or of submicroscopic lymph-node metastases in breast cancer patients (Merrie et al, 1999; Sabbatini et al, 2000). These new informations concerning maspin overexpression in breast cancer associated with a poor prognosis also call into question the use of this gene or its product to inhibit angiogenesis or metastasis.
In conclusion, the results of this study point to a major role of the maspin gene in ERα-positive breast cancer. In particular, we obtained evidence that maspin mRNA status might serve as a new prognostic marker, and also as a possible predictor of the response to endocrine therapy in postmenopausal breast cancer patients. Finally, we describe a rapid, highly sensitive, high-throughput RT–PCR assay for determining maspin mRNA status.
Change history
16 November 2011
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Bièche I, Noguès C, Lidereau R (1999a) Overexpression of BRCA2 gene in sporadic breast tumours. Oncogene 18: 5232–5238
Bièche I, Onody P, Laurendeau I, Olivi M, Vidaud D, Lidereau R, Vidaud M (1999b) Real-time reverse transcription – PCR assay for future management of ERBB2-based clinical applications. Clin Chem 45: 1148–1156
Bièche I, Parfait B, Laurendeau I, Girault I, Vidaud M, Lidereau R (2001) Quantitation of estrogen receptor α and β expression in sporadic breast cancer. Oncogene 20: 8109–8115
Cox DR (1972) Regression models and life-tables. J R Stat Soc (B) 34: 187–220
Domann FE, Rice JC, Hendrix MJ, Futscher BW (2000) Epigenetic silencing of maspin gene expression in human breast cancers. Int J Cancer 85: 805–810
Hojo T, Akiyama Y, Nagasaki K, Maruyama K, Kikuchi K, Ikeda T, Kitajima M, Yamaguchi K (2001) Association of maspin expression with the malignancy grade and tumor vascularization in breast cancer tissues. Cancer Lett 171: 103–110
Jiang WG, Hiscox S, Horrobin DF, Bryce RP, Mansel RE (1997) γ Linolenic acid regulates expression of maspin and the motility of cancer cells. Biochem Biophys Res Commun 237: 639–644
Jiang N, Meng Y, Zhang S, Mensah-Osman E, Sheng S (2002) Maspin sensitizes breast carcinoma cells to induced apoptosis. Oncogene 21: 4089–4098
Kaplan EL, Meier P (1958) Nonparametric estimation from incomplete observations. J Am Stat Assoc 53: 457–481
Maass N, Hojo T, Rosel F, Ikeda T, Jonat W, Nagasaki K (2001a) Down regulation of the tumor suppressor gene maspin in breast carcinoma is associated with a higher risk of distant metastasis. Clin Biochem 34: 303–307
Maass N, Hojo T, Ueding M, Lüttges J, Klöppel G, Jonat W, Nagasaki K (2001c) Expression of the tumor suppressor gene maspin in human pancreatic cancers. Clin Cancer Res 7: 812–817
Maass N, Teffner M, Rosel F, Pawaresch R, Jonat W, Nagasaki K, Rudolph P (2001b) Decline in the expression of the serine proteinase inhibitor maspin is associated with tumour progression in ductal carcinomas of the breast. J Pathol 195: 321–326
Martin KJ, Kritzman BM, Price LM, Koh B, Kwan C-P, Zhang X, Mackay A, O'Hare MJ, Kaelin CM, Mutter GL, Pardee AB, Sager R (2000) Linking gene expression patterns to therapeutic groups in breast cancer. Cancer Res 60: 2232–2238
Merrie AE, Yun K, Gunn J, Phillips LV, Mc Call JL (1999) Analysis of potential markers for detection of submicroscopic lymph node metastases in breast cancer. Br J Cancer 80: 2019–2024
Pemberton PA, Tipton AR, Pavloff N, Smith J, Erickson JR, Mouchabeck ZM, Kiefer MC (1997) Maspin is an intracellular serpin that partitions into secretory vesicles and is present at the cell surface. J Histochem Cytochem 45: 1697–1706
Peto R, Pike MC, Armitage P (1977) Design and analysis of randomized clinical trials requiring prolonged observation of each patient. II. Analysis and examples. Br J Cancer 35: 1–39
Sabbatini R, Frederico M, Morselli M, Depenni R, Cagossi K, Luppi M, Torelli G, Silingardi V (2000) Detection of circulating tumor cells by reverse transcriptase polymerase chain reaction of maspin in patients with breast cancer undergoing conventional-dose chemotherapy. J Clin Oncol 18: 1914–1920
Seftor REB, Seftor EA, Sheng S, Pemberton PA, Sager R, Hendrix MJC (1998) Maspin suppresses the invasive phenotype of human breast carcinoma. Cancer Res 58: 5681–5685
Sheng S, Carey J, Seftor EA, Dias L, Hendrix MJC, Sager R (1996) Maspin acts at the cell membrane to inhibit invasion and motility of mammary and prostatic cancer cells. Proc Natl Acad Sci USA 93: 11669–11674
Sheng S, Truong B, Fredrickson D, Wu R, Pardee AB, Sager R (1998) Tissue-type plasminogen activator is a target of the tumor suppressor gene maspin. Proc Natl Acad Sci USA 95: 499–504
Shi HY, Zhang W, Liang R, Abraham S, Kittrell FS, Medina D, Zhang M (2001) Blocking tumor growth, invasion, and metastasis by maspin in syngeneic breast cancer model. Cancer Res 61: 6945–6951
Speirs V, Malone C, Walton DS, Kerin MJ, Atkin SL (1999) Increased expression of estrogen receptor beta mRNA in tamoxifen-resistant breast cancer patients. Cancer Res 59: 5421–5424
Stal O, Borg A, Fernö M, Kallström A-C, Malmström P, Nordenskjöld B and members of the South Sweden Breast Cancer Group and the Southeast Sweden Breast Cancer Group (2000) ErbB2 status and the benefit from two of five years of adjuvant tamoxifen in postmenopausal early stage breast cancer. Ann Oncol 11: 1545–1550
Stephens RW, Brunner N, Janicke F, Schmitt M (1998) The urokinase plasminogen activator system as a target for prognostic studies in breast cancer. Breast Cancer Res Treat 52: 99–111
Sternlicht MD, Barsky SH (1997) The myoepithelial defense: a host defense against cancer. Med Hypotheses 48: 37–46
Streuli CH (2002) Maspin is a tumour supressor that inhibits breast cancer tumour metastasis in vivo. Breast Cancer Res 4: 137–140
Umerika Y, Ohi Y, Sagara Y, Yoshida H (2002) Expression of maspin predicts poor prognosis in breast cancer patients. Int J Cancer 100: 452–455
Zhang M, Maass N, Magit D, Sager R (1997) Transactivation through Ets and Ap1 transcriptional sites determines the expression of the tumor-suppressing gene maspin. Cell Growth Differ 8: 179–186
Zhang M, Shi Y, Magit D, Furth PA, Sager R (2000b) Reduced mammary tumor progression in WAP-TAg/WAP-maspin bitransgenic mice. Oncogene 19: 6053–6058
Zhang M, Volpert O, Shi YH, Bouck N (2000a) Maspin is an angiogenesis inhibitor. Nat Med 6: 196–1999
Zou Z, Anisowicz A, Hendrix MJC, Thor A, Neveu M, Sheng S, Rafidi K, Seftor E, Sager R (1994) Maspin, a serpin with tumor-suppressing activity in human mammary epithelial cells. Science 263: 526–529
Zou Z, Gao C, Nagaich AK, Connell T, Saito S, Moul JW, Seth P, Appella E, Srivastava S (2000) P53 regulates the expression of the tumor suppressor gene maspin. J Biol Chem 275: 6051–6054
Acknowledgements
This work was supported by the Comité Régional des Hauts-de-Seine de la Ligue Nationale Contre le Cancer. We also thank the staff of Centre René Huguenin for assistance in specimen collection and patient care. We thank Odile Champy for excellent technical assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Bièche, I., Girault, I., Sabourin, JC. et al. Prognostic value of maspin mRNA expression in ERα-positive postmenopausal breast carcinomas. Br J Cancer 88, 863–870 (2003). https://doi.org/10.1038/sj.bjc.6600812
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.bjc.6600812
Keywords
This article is cited by
-
Association between SNPs in Serpin gene family and risk of esophageal squamous cell carcinoma
Tumor Biology (2015)
-
Nuclear location of tumor suppressor protein maspin inhibits proliferation of breast cancer cells without affecting proliferation of normal epithelial cells
BMC Cancer (2014)
-
Expression and localization of maspin in cervical cancer and its role in tumor progression and lymphangiogenesis
Archives of Gynecology and Obstetrics (2014)
-
Maspin: molecular mechanisms and therapeutic implications
Cancer and Metastasis Reviews (2012)
-
Maspin expression in normal lung and non-small-cell lung cancers: cellular property-associated expression under the control of promoter DNA methylation
Oncogene (2004)