Abstract
The immunoglobulin VH gene mutation status can divide B-cell chronic lymphocytic leukaemia (CLL) into two entities with a different clinical course. Cases with unmutated VH genes, considered to evolve from pregerminal centre (GC) cells, have a worse outcome compared to cases showing mutated VH genes, that is, post-GC derived. Also, telomere length has been reported to be of prognostic significance in CLL. Interestingly, telomerase becomes activated during the GC reaction and an elongation of the telomeres occurs in GC B cells. We performed telomere length and VH gene analysis in a series of 61 CLL cases, in order to investigate if the unique telomere lengthening shown in GC B cells could reflect the telomere status in the two subsets of mutated and unmutated CLL. A novel association was found between VH gene mutation status and telomere length, since significantly shorter telomeres were demonstrated in the unmutated group compared to the mutated group (mean length 4.3 vs 6.3 kbp). Shorter telomeres also constituted a subgroup with a worse prognosis than cases with longer telomeres (median survival 59 vs 159 months). Furthermore, the Ig gene sequence data revealed that samples with high mutations frequency (>6%) had long telomeres (∼8 kbp). Thus, both the telomere and VH gene mutation status in CLL appear linked, which may reflect the proliferative history of the clonal cells with regard to the GC reaction.
Similar content being viewed by others
Main
B-cell chronic lymphocytic leukaemia (CLL) is characterised by a monoclonal expansion of small B lymphocytes, typically expressing CD5, CD19, CD23 and low levels of surface immunoglobulin (Ig) (Matutes et al, 1994), which previously were supposed to originate from naive B cells of the mantle zone. However, recent analyses of the Ig heavy-chain variable region (IgVH) gene have defined two subsets of CLL, comprising cases with somatically unmutated or mutated VH genes (Fais et al, 1998; Hamblin et al, 1999; Damle et al, 1999). The unmutated cases are considered to originate from pregerminal centre (GC) B cells and the mutated from post-GC B cells. A more favourable prognosis has been shown for CLL cases with somatically mutated VH genes compared with unmutated cases (Hamblin et al, 1999; Damle et al, 1999; Maloum et al, 2000; Thunberg et al, 2001), indicating that CLL can be separated into at least two entities with a different clinical outcome.
Human telomeres in normal somatic cells consist of 6–12 kbp of TTAGGG-repeats that are eroded upon cell division because of the so-called ‘end-replication problem’ (Moyzis et al, 1988; Blackburn, 1991; Levy et al, 1992). A decrease in telomere length has been demonstrated with cell division in vitro and with cellular age in vivo for different cell types and tissues. At a critical telomere length during shortening, an ageing programme is activated in normal cells (Allsopp et al, 1992, 1995a, 1995b). Thus, the telomere length can predict and limit the number of divisions a cell can undergo. This telomere erosion, however, can be counteracted by the telomerase complex, which has a reverse transcriptase activity and can synthesise new telomeric repeats (Morin, 1989). Telomerase activity is linked to cells with an extended or infinite division potential, such as stem cells, germline cells and lymphocytes as well as permanent cell lines and tumor cells, while telomerase activity is absent from most normal somatic cells (Kim et al, 1994; Hiyama et al, 1995; Härle-Bachor and Boukamp, 1996; Norrback et al, 1996; Norrback and Roos, 1997). Recently, high levels of telomerase activity have been demonstrated in normal GC B cells in association with a unique telomere lengthening process not demonstrated in other cell types in vivo (Hu et al, 1997; Weng et al, 1997; Norrback et al, 2001).
Different levels of telomerase activity are expressed in the vast majority of leukaemias and malignant lymphomas (Norrback and Roos, 1997). In acute myeloid leukaemia, relapse cases demonstrate higher telomerase activity levels compared to that at diagnosis and similar findings have been reported for advanced CLL cases (Counter et al, 1995; Ohyashiki et al, 1997; Xu et al, 1998). These data indicate that the expression of telomerase activity can be associated with tumour progression in haematological neoplasms. Regarding CLL, one study has shown that both the telomere length and telomerase activity level were significant prognostic markers, since a short median survival was significantly associated with short telomere length (<6 kbp) and high telo-merase activity (Bechter et al, 1998). If the telomere lengthening in GC B cells demonstrated in benign tissues, like tonsils, is of relevance in this context, CLL cases of post-GC type should then be expected to have longer telomeres than pre-GC CLL.
In the present study, we have analysed a series of CLL cases for VH mutation status by sequencing of the clonal rearrangements, for telomere length by Southern blotting and for clinical outcome. We have found an association between VH mutation status and telomere length with shorter telomeres in unmutated (pre-GC) CLL cases compared with mutated (post-GC) CLL. Furthermore, a relationship between VH mutation and telomere length was demonstrated since a gradual increase in telomere length was shown to parallel a decrease in homology to VH germline sequences, that is, an increase in mutation rate.
Materials and methods
Patients
Frozen tumour samples from 61 patients were studied, which were identified in the archives of the departments of Pathology at Uppsala University Hospital and Umeå University Hospital between 1981 and 1998. The tumour material was obtained from bone marrow (29 cases), peripheral blood (26 cases), spleen (five cases) and lymph node (one case). There were 43 men and 18 women. Survival data were available for 56 patients from the local Swedish population and cancer registries in Uppsala and Umeå. The median age at diagnosis was 65 years and the median survival time was 82 months. Patient follow-up ranged from 8 to 182 months with a median follow-up of 56 months. The present study was approved by the ethical committee.
Morphology and immunophenotyping
Classification was performed on smears, sections, imprints and by immunophenotyping using flow cytometry. According to the Royal Marsden scoring system the tumour cells expressed CD5, CD19 and CD23, and a weak expression of Ig (Matutes et al, 1994).
PCR amplification and nucleotide sequencing analysis
High molecular weight DNA was prepared from frozen tumour material using standard protocols including proteinase K treatment, chloroform treatment and ethanol precipitation. VH gene family-specific PCR amplification was performed using six family- specific VH primers and one JH primer as previously described (Li et al, 1999). Monoclonal PCR products were distinguished from polyclonal by a single-strand conformation polymorphism analysis using polyacrylamide gel or GenePhor electrophoresis according to the manufacturer's protocol (Amersham Pharmacia biotech, Uppsala, Sweden).
The clonal PCR products were sequenced directly using the BigDye Terminator Cycle Sequencing Reaction Kit (Perkin-Elmer, ABI, FosterCity, CA, USA) or using cloning as previously described (Li et al, 1999). All sequence reactions were analysed using an automated DNA sequencer (ABI 377, Applied Biosystems, Foster City, CA, USA).
Analysis of VH, D and JH sequences
The sequences were aligned to IgH sequences from the BLAST database (National Center for Biotechnology Information, USA), the V-BASE database (MRC, Centre for Protein Engineering, Cambridge, UK) and the Immunogenetics database (http://imgt.cines.fr:8104, initiator and coordinator: Marie-Paule Lefranc, Montpellier, France). A VH gene sequence deviating more than 2% from the corresponding germline gene was defined as mutated (Matsuda et al, 1993).
Southern blotting
Southern blotting and hybridisation with the telomeric probe (TTAGGG)4 were performed and mean telomere restriction fragment (TRF) length was calculated as previously described (Mehle et al, 1994). As molecular weight standards, the lambda DNA/Eco1 Styl/Mlu1 Marker (MBI Fermentas Inc., Amherst, NY, USA) and the DNA molecular weight marker X (Boehringer Mannheim Gmbh, Germany) were used. The peak TRF value was estimated as the length corresponding to the point with the highest optical density within the TRF profile.
hTERT mRNA expression
In 19 cases with material available for RNA extraction, hTERT mRNA levels were analysed as previously described (Norrback et al, 2001) using a real-time polymerase chain reaction. The amount of hTERT mRNA was quantified using the Light Cycler Telo TAGGG hTERT Quantification kit where hTERT RNA levels were expressed as a ratio between the expression level of hTERT RNA and a house-keeping gene RNA (porphobilinogen deaminase) according to the manufacturer's protocol (Roche, Basel, Switzerland).
Statistical analyses
Kaplan–Meier survival curves and the log-rank test were performed to study the prognostic significance of VH gene mutations and telomere length in CLL using the Statistica 5.5 software (Stat Soft Inc., Tulsa, USA). Survival was calculated from the date of diagnosis until the last follow-up or death. Independent samples t-test was used to compare means, and nonparametric correlation was calculated according to the Spearman rank correlation. Probabilities of less than 0.05 were accepted as a significant value.
Results
Analysis of VH gene mutations
A total of 34 cases (56%) demonstrated unmutated VH genes and 27 cases (44%) somatically mutated VH genes. In the mutated cases, the mutation frequency ranged from 2.1–11.7%. Two clonal rearrangements were found in six cases, five of which showed two unmutated VH genes, whereas one case had one unmutated and one mutated (97.7% homology) VH gene. The last case was considered as mutated and the first five cases as unmutated.
Telomere length and Ig gene mutation status
A summary of the data is given in Table 1. The mean peak TRF length was 5.16 kbp and the median peak TRF length was 4.57 kbp, range 2.35–10.10 kbp. The reason for using peak TRF values in the further analysis was that 10 cases had two peaks and for these the value of the largest peak was used, since all cases had >50% monoclonal B cells as determined by immunophenotyping. The cases with double peaks had only one clonal Ig gene rearrangement.
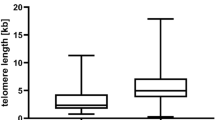
A total of 34 unmutated and 27 mutated samples were analysed for telomere length. The unmutated cases demonstrated a median peak TRF length of 4.24 kbp (mean peak 4.29 kbp, range 2.35–7.66 kbp) compared with 5.69 kbp (mean peak 6.25 kbp, range 2.66–10.10 kbp) for the mutated cases. The association between mutation status and telomere length (Figure 1) was significant using the Spearman rank correlation (R2=0.46, P<0.001).
Thus, the IgVH gene mutation status correlated with TRF length. For cases with no somatic hypermutations (i.e. 100% homology to the germline gene), the TRF length was ∼4 kpb and for cases with a high mutation rate (<94% homology to the germline gene), the TRF length was ∼8 kbp. The material was divided into two groups of equal size with a cutoff at the median peak TRF value (4.57 kbp).
The presence of hTERT RNA was detected in all the 19 cases studied. Using independent samples' t-test, a significant association (P=0.032) was found between long telomeres (TRF >4.57 kbp) and low hTERT mRNA levels and vice versa (data not shown). No association was found between mutation status and hTERT mRNA levels.
Survival analysis
Survival curves were plotted according to the Kaplan–Meier method using data available on 56 patients. The survival data were grouped into mutated (26 patients) and unmutated (30 patients) cases and a statistically significant difference in overall survival was found between these two groups (P<0.001) (Figure 2). The median survival for patients with mutated VH genes was 138 months and for those without mutations 55 months, calculated from the date of diagnosis. The median age was 63 years for patients with unmutated VH genes and 67 years for patients with mutated VH genes.
Regarding telomere length, survival analysis was performed after dividing the material into two groups with a cutoff at the median peak TRF value (4.57 kbp). A significant difference in survival was found between the two groups with a better overall survival for cases with peak TRF values >4.57 kbp (P=0.0015) (Figure 3). The median survival for patients with telomeres longer than the cutoff value was 159 months and for those with shorter telomeres 59 months. The median age was 62 years for patients with TRF lengths <4.57 kbp and 67 years for cases with TRF lengths >4.57 kbp. Using Cox regression analysis neither mutation status nor telomere length were independent variables, indicating an association between these two parameters.
Discussion
Convincing data have made it probable that CLL consists of at least two variants derived from different subpopulations of neoplastic cells (Fais et al, 1998; Damle et al, 1999; Hamblin et al, 1999; Maloum et al, 2000), one originating from unmutated pre-GC cells and the other from somatically hypermutated GC or post-GC cells. This notion was further supported by the observation that patients with mutated VH genes had a significantly better outcome compared to patients lacking VH gene mutations (Damle et al, 1999; Hamblin et al, 1999; Maloum et al, 2000; Thunberg et al, 2001). In the present study, we were able to verify these data regarding prognosis and VH gene mutation status, since our mutated CLL cases displayed more than twice as long median survival than the unmutated cases (138 vs 55 months). Most interestingly, we found a novel association between VH gene mutation frequency and telomere length, showing significantly longer telomeres in CLL cases having somatically mutated VH genes. We could also demonstrate that CLL cases with longer telomeres had a significantly better prognosis than cases with shorter telomeres (159 vs 59 months), supporting a previously published study (Bechter et al, 1998). But what could be the biological explanation for this new association coupling together our findings?
Normal lymphocytes have been shown to lose telomeres with each cell division in vivo and in vitro (Vaziri et al, 1994; Weng et al, 1995; Norrback and Roos, 1997; Rufer et al, 1998). However, GC B cells are characterised by telomerase activity and a unique telomere lengthening process has been demonstrated in crude preparations of GC B cells (Hu et al, 1997; Weng et al, 1997; Norrback et al, 2001). In immunopurified cell populations from tonsils, we found a telomere lengthening in both the centroblast and centrocyte populations (Norrback et al, 2001). The telomere length increase is likely to occur in proliferating pre-GC blasts and centroblasts, since telomerase activity is coupled to cell cycle progression in lymphocytes (Buchkovich and Greider, 1996; Igarashi and Sakaguchi, 1997). In cells maturing from naive B cells to centroblasts/centrocytes, we could therefore demonstrate a telomere lengthening of up to 4 kbp in individual cases (Norrback et al, 2001).
In our CLL material, mutated cases showed roughly 2 kb longer telomeres than the unmutated. We therefore believe that this difference in telomere length between mutated and unmutated CLL reflects, and supports the theory, that these subsets probably originate from different stages in the B-cell development, that is, pre- and post-GC B cells. Interestingly, the difference in telomere length between pre-GC and post-GC cells seemed to be preserved in the CLL cases, which may indicate a similar rate of telomere loss in the different CLL clones. The shorter telomere lengths in unmutated and mutated CLL compared to normal pre-GC and GC B cells (Norrback et al, 2001) could, at least partly, be explained by telomere loss normally occurring during ageing, since most of the normal GC B cells studied by us were derived from tonsils from young individuals whereas the CLL cases represented an older patient group.
Telomerase is activated in the vast majority of leukaemias and malignant lymphomas, including CLL (reviewed in Norrback and Roos, 1997). In a subset of our CLL cohort (n=19), we have analysed the mRNA levels of the catalytic unit of telomerase, hTERT, which have been shown to correlate with the telomerase activity in B lymphocytes (Liu et al, 1999; Norrback et al, 2001). We found that CLL cases with longer telomeres had lower hTERT mRNA levels than cases with shorter telomeres, which is in line with a previously published report on telomerase activity (Bechter et al, 1998). These collected data argue that the difference in telomere lengths between the two groups seemed unrelated to hTERT RNA levels (and thus telomerase activity), and rather reflected the telomere status of the primarily transformed cells.
One common denominator for telomerase activation and the VH gene hypermutation mechanism in the GC is an active cell cycle. Somatic VH gene hypermutations occur in growing B cells given the survival signals based on antigen selection, and in these cells telomerase is known to be upregulated as discussed above. The combined telomere length and VH gene hypermutation analysis presented here might give new insights into the kinetics of the hypermutation machinery. It can be anticipated that the total telomere length increase is a result of the number of cell cycle rounds undergone in the GC. Thus, the data suggest that a limited number of VH gene mutations occur for each GC cell division round since post-GC B cells with long telomeres had high VH gene mutation frequencies. This association between VH gene mutation status and telomere length provides new possibilities for more detailed analyses of GC B cell subpopulations. It should be noted that an alternative way to acquire somatic VH gene mutations has been described in the hyper-IgM syndrome because of a nonfunctional CD40–CD154 interaction leading to no development of germinal centres (Durandy and Honjo, 2001). However, the mutation frequency in the hyper-IgM syndrome (usually <2%) (Chu et al, 1995; Monson et al, 2001; Weller et al, 2001) is below the cutoff level used to distinguish between mutated and unmutated CLL cases. Hence, it seems unlikely that the mutated CLL cases had acquired VH gene mutations by the alternative CD40-independent pathway.
Finally, the survival data also suggest that telomere length can be an important clinical parameter to study for the evaluation of CLL in the diagnostic setting, since longer telomere lengths were associated with better clinical outcome. We have chosen the median value as cutoff level, but the true border to divide the material into prognostic groups deserves further investigation. Recently described techniques for flow cytometric analysis after in situ hybridisation with a fluorochrome labelled telomere probe (‘flow-FISH’) can be useful (Rufer et al, 1998; Hultdin et al, 1998), since the flow-FISH approach is convenient and can be directly applied to material sent for regular diagnostic work. However, further studies of larger groups of patients have to be performed to clarify the clinical impact of telomere length analysis in CLL.
Change history
16 November 2011
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Allsopp RC, Chang E, Kashefi-Aazam M, Rogaev EI, Piatyszek MA, Shay JW, Harley CB (1995a) Telomere shortening is associated with cell division in vitro and in vivo. Exp Cell Res 220: 194–200
Allsopp RC, Harley CB (1995b) Evidence for a critical telomere length in senescent human fibroblasts. Exp Cell Res 219: 130–136
Allsopp RC, Vaziri H, Patterson C, Goldstein S, Younglai EV, Futcher AB, Greider CW, Harley CB (1992) Telomere length predicts replicative capacity of human fibroblasts. Proc Natl Acad Sci USA 89: 10114–10118
Bechter OE, Eisterer W, Pall G, Hilbe W, Kühr T, Thaler J (1998) Telomere length and telomerase activity predict survival in patients with B cell chronic lymphocytic leukemia. Cancer Res 58: 4918–4922
Blackburn EH (1991) Structure and function of telomeres. Nature 350: 569–572.
Buchkovich KJ, Greider CW (1996) Telomerase regulation during entry into the cell cycle in normal human T cells. Mol Biol Cell 7: 1443–1454
Chu YW, Marin E, Fuleihan R, Ramesh N, Rosen FS, Geha RS, Insel RA (1995) Somatic mutation of human immunoglobulin V genes in the X-linked HyperIgM syndrome. J Clin Invest 95: 1389–1393
Counter CM, Gupta J, Harley CB, Leber B, Bacchetti S (1995) Telomerase activity in normal leukocytes and in hematologic malignancies. Blood 85: 2315–2320
Damle RN, Wasil T, Fais F, Ghiotto F, Valetto A, Allen SL, Buchbinder A, Budman D, Dittmar K, Kolitz J, Lichtman SM, Schulman P, Vinciguerra VP, Rai KR, Ferrarini M, Chiorazzi N (1999) Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood 94: 1840–1847
Durandy A, Honjo T (2001) Human genetic defects in class-switch recombination (hyper-IgM syndromes). Curr Opin Immunol 13: 543–548
Fais F, Ghiotto F, Hashimoto S, Sellars B, Valetto A, Allen SL, Schulman P, Vinciguerra VP, Rai K, Rassenti LZ, Kipps TJ, Dighiero G, Schroeder H, Ferrarini M, Chiorazzi N (1998) Chronic lymphocytic leukemia B cells express restricted sets of mutated and unmutated antigen receptors. J Clin Invest 102: 1515–1525
Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK (1999) Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood 94: 1848–1854
Härle-Bachor C, Boukamp P (1996) Telomerase activity in the regenerative basal layer of the epidermis in human skin and in immortal and carcinoma-derived skin keratinocytes. Proc Natl Acad Sci USA 93: 6476–6481
Hiyama K, Hirai Y, Kyoizumi S, Akiyama M, Hiyama E, Piatyszek MA, Shay JW, Ishioka S, Yamakido M (1995) Activation of telomerase in human lymphocytes and hematopoietic progenitor cells. J Immunol 155: 3711–3715
Hu BT, Lee SC, Marin E, Ryan DH, Insel RA (1997) Telomerase is up-regulated in human germinal center B cells in vivo and can be re-expressed in memory B cells activated in vitro. J Immunol 159: 1068–1071
Hultdin M, Grönlund E, Eriksson-Lindström E, Norrback K-F, Just T, Roos G (1998) Telomere analysis by fluorescence in situ hybridization and flow cytometry. Nucleic Acids Res 26: 3651–3656
Igarashi H, Sakaguchi N (1997) Telomerase activity is induced in human peripheral B lymphocytes by the stimulation to antigen receptor. Blood 89: 1299–1307
Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PLC, Coviello GM, Wright WE, Weinrich SL, Shay JW (1994) Specific association of human telomerase activity with immortal cells and cancer. Science 266: 2011–2015
Levy MZ, Allsopp RC, Futcher AB, Greider CW, Harley CB (1992) Telomere end-replication problem and cell ageing. J Mol Biol 225: 951–960
Li AH, Rosenquist R, Forestier E, Holmberg D, Lindh J, Löfvenberg E, Roos G (1999) Clonal rearrangements in childhood and adult precursor B acute lymphoblastic leukemia: a comparative polymerase chain reaction study using multiple sets of primers. Eur J Haematol 63: 211–218
Liu K, Schoonmaker MM, Levine BL, June CH, Hodes RJ, Weng N-P (1999) Constitutive and regulated expression of telomerase reverse transcriptase (hTERT) in human lymphocytes. Proc Natl Acad Sci USA 96: 5147–5152
Maloum K, Davi F, Merle-Beral H, Pritsch O, Magnac C, Vuillier F, Dighiero G, Troussard X, Mauro FF, Benichou J (2000) Expression of unmutated VH genes is a detrimental prognostic factor in chronic lymphocytic leukemia. Blood 96: 377–379
Matsuda F, Shin EK, Nagaoka H, Matsumura R, Haino M, Fukita Y, Takaishi S, Imai T, Riley JH, Anand R, Soeda E, Honjo T (1993) Structure and physical map of 64 variable segments in the 3'0.8- megabase region of the human immunoglobulin heavy-chain locus. Nat Genet 3: 88–94
Matutes E, Owusu-Ankomah K, Morilla R, Garcia Marco J, Houlihan A, Que TH, Catovsky D (1994) The immunological profile of B-cell disorders and proposal of a scoring system for the diagnosis of CLL. Leukemia 8: 1640–1645
Mehle C, Ljungberg B, Roos G (1994) Telomere shortening in renal cell carcinoma. Cancer Res 54: 236–241
Monson NL, Foster SJ, Brezinschek HP, Brezinschek RI, Dorner T, Lipsky PE (2001) The role of CD40–CD40 ligand (CD154) interactions in immunoglobulin light chain repertoire generation and somatic mutation. Clin Immunol 100: 71–81
Morin GB (1989) The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell 59: 521–529
Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD, Meyne J, Ratliff RL, Wu J-R (1988) A highly conserved repetitive DNA sequence, (TTAGGG), present at the telomeres of human chromosomes. Proc Natl Acad Sci USA 85: 6622–6626
Norrback K-F, Dahlenborg K, Carlsson R, Roos G (1996) Telomerase activation in normal B lymphocytes and Non-Hodgkin's lymphomas. Blood 88: 222–229
Norrback K-F, Hultdin M, Dahlenborg K, Osterman P, Carlsson R, Roos G (2001) Telomerase regulation and telomere dynamics in human germinal centers. Eur J Haematol 67: 309–317
Norrback K-F, Roos G (1997) Telomeres and telomerase in normal and malignant haematopoietic cells. Eur J Cancer 33: 774–780
Ohyashiki JH, Ohyashiki K, Iwama H, Hayashi S, Toyama K, Shay JW (1997) Clinical implications of telomerase activity levels in acute leukemia. Clin Cancer Res 3: 619–625
Rufer N, Dragowska W, Dumont-Girard F, Chapuis B, Roosnek E, Lansdorp PM (1998) Telomere length dynamics in human lymphocyte subpopulations measured by flow cytometry. Nat Biotechnol 16: 743–747
Thunberg U, Johnson A, Roos G, Thorn I, Tobin G, Sallstrom J, Sundstrom C, Rosenquist R (2001) CD38 expression is a poor predictor for VH gene mutational status in chronic lymphocytic leukemia. Blood 97: 1892–1893
Vaziri H, Dragowska W, Allsopp RC, Thomas TE, Harley CB, Lansdorp PM (1994) Evidence for a mitotic clock in human hematopoietic stem cells: loss of telomeric DNA with age. Proc Natl Acad Sci USA 91: 9857–9860
Weller S, Faili A, Garcia C, Braun MC, Le Deist F, de Saint Basile G, Hermine O, Fischer A, Reynaud CA, Weill JC (2001) CD40–CD40L independent Ig gene hypermutation suggests a second B cell diversification pathway in humans. Proc Natl Acad Sci USA 98: 1166–1170
Weng N-P, Granger L, Hodes RJ (1997) Telomere lengthening and telomerase activation during human B cell differentiation. Proc Natl Acad Sci USA 94: 10827–10832
Weng N-P, Levine BL, June CH, Hodes RJ (1995) Human naive and memory T lymphocytes differ in telomeric length and replicative potential. Proc Natl Acad Sci USA 92: 11091–11094
Xu D, Gruber A, Peterson C, Pisa P (1998) Telomerase activity and the expression of telomerase components in acute myelogenous leukaemia. Br J Haematol 102: 1367–1375
Acknowledgements
We are grateful to Eleonor Lindström-Eriksson and Ulla-Britt Westman for their skilful technical assistance. This study was supported by grants from the Swedish Cancer Society, the Medical Faculty, Umeå University, Lion's Cancer Research Foundation at Umeå University and Uppsala University, and by Grant QLG1-1999-01341 from the European Union.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Hultdin, M., Rosenquist, R., Thunberg, U. et al. Association between telomere length and VH gene mutation status in chronic lymphocytic leukaemia: clinical and biological implications. Br J Cancer 88, 593–598 (2003). https://doi.org/10.1038/sj.bjc.6600763
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.bjc.6600763
Keywords
This article is cited by
-
Association between telomere length and survival in cancer patients: a meta-analysis and review of literature
Frontiers of Medicine (2016)
-
Molecular basis of telomere dysfunction in human genetic diseases
Nature Structural & Molecular Biology (2015)
-
Telomeres and prognosis in patients with chronic lymphocytic leukaemia
International Journal of Hematology (2011)
-
Telomere length is an independent predictor of survival, treatment requirement and Richter's syndrome transformation in chronic lymphocytic leukemia
Leukemia (2009)
-
Telomerase and cancer therapeutics
Nature Reviews Cancer (2008)