Abstract
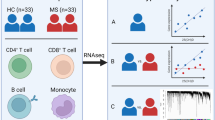
Given the heterogeneous nature of multiple sclerosis (MS), we applied DNA microarray technology to determine whether variability is reflected in peripheral blood (PB) cells. In this study, we studied whole-blood gene expression profiles of 29 patients with relapsing-remitting MS (RRMS) and 25 age- and sex-matched healthy controls. We used microarrays with a complexity of 43K cDNAs. The data were analyzed using sophisticated pathway-level analysis in order to provide insight into the deregulated peripheral immune response programs in MS. We found a remarkable elevated expression of a spectrum of genes known to be involved in immune defense in the PB of MS patients compared to healthy individuals. Cluster analysis revealed that the increased expression of these genes was characteristic for approximately half of the patients. In addition, the gene signature in this group of patients was comparable with a virus response program. We conclude that the transcriptional signature of the PB cells reflects the heterogeneity of MS and defines a sub-population of RRMS patients, who exhibit an activated immune defense program that resembles a virus response program, which is supportive for a link between viruses and MS.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 digital issues and online access to articles
$119.00 per year
only $19.83 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Hafler DA . Multiple sclerosis. J Clin Invest 2004; 113: 788–794.
Rio J, Nos C, Tintore M, Borras C, Galan I, Comabella M et al. Assessment of different treatment failure criteria in a cohort of relapsing-remitting multiple sclerosis patients treated with interferon beta: implications for clinical trials. Ann Neurol 2002; 52: 400–406.
Lucchinetti C, Bruck W, Parisi J, Scheithauer B, Rodriguez M, Lassmann H . Heterogeneity of multiple sclerosis lesions: implications for the pathogenesis of demyelination. Ann Neurol 2000; 47: 707–717.
Bielekova B, Kadom N, Fisher E, Jeffries N, Ohayon J, Richert N et al. MRI as a marker for disease heterogeneity in multiple sclerosis. Neurology 2005; 65: 1071–1076.
Sadovnick AD, Armstrong H, Rice GP, Bulman D, Hashimoto L, Paty DW et al. A population-based study of multiple sclerosis in twins: update. Ann Neurol 1993; 33: 281–285.
Pugliatti M, Sotgiu S, Rosati G . The worldwide prevalence of multiple sclerosis. Clin Neurol Neurosurg 2002; 104: 182–191.
Kurtzke JF . Epidemiology of MS. In: Hallpike JF, Adams CWM, Tourtellote WE (eds). Multiple Sclerosis. Willimas and Wilkins: Baltimore, MD, 2005, pp. 49–95.
Ter Meulen V, Katz M . The proposed viral etiology of multiple sclerosis and related demyelinating diseases. In: Raine CS, MacFarland HF, Tourtellotte WW (eds). Multiple Sclerosis. Chapman & Hall: London, 1997, pp. 287–305.
Tusher VG, Tibshirani R, Chu G . Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 2001; 98: 5116–5121.
Cho RJ, Campbell MJ . Transcription, genomes, function. Trends Genet 2000; 16: 409–415.
Mi H, Lazareva-Ulitsky B, Loo R, Kejariwal A, Vandergriff J, Rabkin S et al. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res 2005; 33: D284–D288.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102: 15545–15550.
Jenner RG, Young RA . Insights into host responses against pathogens from transcriptional profiling. Nat Rev Microbiol 2005; 3: 281–294.
Samuel CE . Antiviral actions of interferons. Clin Microbiol Rev 2001; 14: 778–809, Table.
Moynagh PN . TLR signalling and activation of IRFs: revisiting old friends from the NF-kappaB pathway. Trends Immunol 2005; 26: 469–476.
Rubins KH, Hensley LE, Jahrling PB, Whitney AR, Geisbert TW, Huggins JW et al. The host response to smallpox: analysis of the gene expression program in peripheral blood cells in a nonhuman primate model. Proc Natl Acad Sci USA 2004; 101: 15190–15195.
Achiron A, Gurevich M, Friedman N, Kaminski N, Mandel M . Blood transcriptional signatures of multiple sclerosis: unique gene expression of disease activity. Ann Neurol 2004; 55: 410–417.
Aune TM, Maas K, Moore JH, Olsen NJ . Gene expression profiles in human autoimmune disease. Curr Pharm Des 2003; 9: 1905–1917.
Maas K, Chan S, Parker J, Slater A, Moore J, Olsen N et al. Cutting edge: molecular portrait of human autoimmune disease. J Immunol 2002; 169: 5–9.
Mandel M, Gurevich M, Pauzner R, Kaminski N, Achiron A . Autoimmunity gene expression portrait: specific signature that intersects or differentiates between multiple sclerosis and systemic lupus erythematosus. Clin Exp Immunol 2004; 138: 164–170.
Satoh J, Nakanishi M, Koike F, Miyake S, Yamamoto T, Kawai M et al. Microarray analysis identifies an aberrant expression of apoptosis and DNA damage-regulatory genes in multiple sclerosis. Neurobiol Dis 2005; 18: 537–550.
Valyi-Nagy T, Dermody TS . Role of oxidative damage in the pathogenesis of viral infections of the nervous system. Histol Histopathol 2005; 20: 957–967.
Hultqvist M, Olofsson P, Holmberg J, Backstrom BT, Tordsson J, Holmdahl R . Enhanced autoimmunity, arthritis, and encephalomyelitis in mice with a reduced oxidative burst due to a mutation in the Ncf1 gene. Proc Natl Acad Sci USA 2004; 101: 12646–12651.
Patterson JB, Thomis DC, Hans SL, Samuel CE . Mechanism of interferon action: double-stranded RNA-specific adenosine deaminase from human cells is inducible by alpha and gamma interferons. Virology 1995; 210: 508–511.
Lomeli H, Mosbacher J, Melcher T, Hoger T, Geiger JR, Kuner T et al. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science 1994; 266: 1709–1713.
Player MR, Torrence PF . The 2–5A system: modulation of viral and cellular processes through acceleration of RNA degradation. Pharmacol Ther 1998; 78: 55–113.
Reichelt M, Stertz S, Krijnse-Locker J, Haller O, Kochs G . Missorting of LaCrosse virus nucleocapsid protein by the interferon-induced MxA GTPase involves smooth ER membranes. Traffic 2004; 5: 772–784.
Stranden AM, Staeheli P, Pavlovic J . Function of the mouse Mx1 protein is inhibited by overexpression of the PB2 protein of influenza virus. Virology 1993; 197: 642–651.
Chieux V, Chehadeh W, Hautecoeur P, Harvey J, Wattre P, Hober D . Increased levels of antiviral MxA protein in peripheral blood of patients with a chronic disease of unknown etiology. J Med Virol 2001; 65: 301–308.
Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Ortmann WA, Espe KJ et al. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci USA 2003; 100: 2610–2615.
van der Pouw Kraan CTM, Wijbrandts CA, van Baarsen LGM, Voskuyl AE, Rustenburg F, Baggen JM et al. Rheumatoid arthritis subtypes identified by genomic profiling of peripheral blood cells: assignement of a type I interferon signature in a subpopulation of patients (submitted for publication).
Bave U, Nordmark G, Lovgren T, Ronnelid J, Cajander S, Eloranta ML et al. Activation of the type I interferon system in primary Sjogren's syndrome: a possible etiopathogenic mechanism. Arthritis Rheum 2005; 52: 1185–1195.
Huang X, Yuang J, Goddard A, Foulis A, James RF, Lernmark A et al. Interferon expression in the pancreases of patients with type I diabetes. Diabetes 1995; 44: 658–664.
Greenberg SA, Pinkus JL, Pinkus GS, Burleson T, Sanoudou D, Tawil R et al. Interferon-alpha/beta-mediated innate immune mechanisms in dermatomyositis. Ann Neurol 2005; 57: 664–678.
Bengtsson AA, Sturfelt G, Truedsson L, Blomberg J, Alm G, Vallin H et al. Activation of type I interferon system in systemic lupus erythematosus correlates with disease activity but not with antiretroviral antibodies. Lupus 2000; 9: 664–671.
Akira S, Takeda K . Toll-like receptor signalling. Nat Rev Immunol 2004; 4: 499–511.
Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol 2004; 5: 730–737.
van Noort JM, Bajramovic JJ, Plomp AC, van Stipdonk MJ . Mistaken self, a novel model that links microbial infections with myelin-directed autoimmunity in multiple sclerosis. J Neuroimmunol 2000; 105: 46–57.
Lovgren T, Eloranta ML, Bave U, Alm GV, Ronnblom L . Induction of interferon-alpha production in plasmacytoid dendritic cells by immune complexes containing nucleic acid released by necrotic or late apoptotic cells and lupus IgG. Arthritis Rheum 2004; 50: 1861–1872.
Weinstock-Guttman B, Badgett D, Patrick K, Hartrich L, Santos R, Hall D et al. Genomic effects of IFN-beta in multiple sclerosis patients. J Immunol 2003; 171: 2694–2702.
Koike F, Satoh J, Miyake S, Yamamoto T, Kawai M, Kikuchi S et al. Microarray analysis identifies interferon beta-regulated genes in multiple sclerosis. J Neuroimmunol 2003; 139: 109–118.
Baranzini SE, Mousavi P, Rio J, Caillier SJ, Stillman A, Villoslada P et al. Transcription-based prediction of response to IFNbeta using supervised computational methods. PLoS Biol 2005; 3: e2.
Sturzebecher S, Wandinger KP, Rosenwald A, Sathyamoorthy M, Tzou A, Mattar P et al. Expression profiling identifies responder and non-responder phenotypes to interferon-beta in multiple sclerosis. Brain 2003; 126: 1419–1429.
McDonald WI, Compston A, Edan G, Goodkin D, Hartung HP, Lublin FD et al. Recommended diagnostic criteria for multiple sclerosis: guidelines from the International Panel on the diagnosis of multiple sclerosis. Ann Neurol 2001; 50: 121–127.
van der Pouw Kraan TC, Baarsen EGM, Rustenburg F et al. Gene expression profiling in rheumatology. In: Cope A (ed). Methods in Molecular Medicine. The Humana Press Inc.: Totowa, NJ, 2005.
Van Gelder RN, von Zastrow ME, Yool A, Dement WC, Barchas JD, Eberwine JH . Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc Natl Acad Sci USA 1990; 87: 1663–1667.
Perou CM, Jeffrey SS, van de RM, Rees CA, Eisen MB, Ross DT et al. Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proc Natl Acad Sci USA 1999; 96: 9212–9217.
Ball CA, Awad IA, Demeter J, Gollub J, Hebert JM, Hernandez-Boussard T et al. The standard microarray database accommodates additional microarray platforms and data formats. Nucleic Acids Res 2005; 33 (Database issue): D580–D582.
Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J et al. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res 2002; 30: e15.
Eisen MB, Spellman PT, Brown PO, Botstein D . Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 1998; 95: 14863–14868.
Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res 2003; 13: 2129–2141.
Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 2003; 34: 267–273.
Acknowledgements
This study was partly supported by the Centre for Medical Systems Biology (a center of excellence approved by the Netherlands Genomics Initiative/Netherlands Organization for Scientific Research), and grants from the Stichting MS Research. We would like to thank the following authors for making their microarray data publicly available: RG Jenner (Cambridge, USA), KH Rubins (Stanford, USA) and Q Huang (Cambridge, USA).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on Genes and Immunity website (http://www.nature.com/gene)
Supplementary information
Rights and permissions
About this article
Cite this article
van Baarsen, L., van der Pouw Kraan, T., Kragt, J. et al. A subtype of multiple sclerosis defined by an activated immune defense program. Genes Immun 7, 522–531 (2006). https://doi.org/10.1038/sj.gene.6364324
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.gene.6364324
Keywords
This article is cited by
-
The link between circulating follicular helper T cells and autoimmunity
Nature Reviews Immunology (2022)
-
Association of SHMT1, MAZ, ERG, and L3MBTL3 Gene Polymorphisms with Susceptibility to Multiple Sclerosis
Biochemical Genetics (2019)
-
Predictors of Response to Multiple Sclerosis Therapeutics in Individual Patients
Drugs (2016)
-
Physiological evidence for diversification of IFNα- and IFNβ-mediated response programs in different autoimmune diseases
Arthritis Research & Therapy (2016)
-
Elevated type I interferon-like activity in a subset of multiple sclerosis patients: molecular basis and clinical relevance
Journal of Neuroinflammation (2012)