Abstract
Mitochondrial DNA (mtDNA) mutations have been implicated in non-syndromic hearing loss either as primary or as predisposing factors. As only a part of the mitochondrial genome is usually explored in deafness, its prevalence is probably under-estimated. Among 1350 families with non-syndromic sensorineural hearing loss collected through a French collaborative network, we selected 29 large families with a clear maternal lineage and screened them for known mtDNA mutations in 12S rRNA, tRNASer(UCN) and tRNALeu(UUR) genes. When no mutation could be identified, a whole mitochondrial genome screening was performed, using a microarray resequencing chip: the MitoChip version 2.0 developed by Affymetrix Inc. Known mtDNA mutations was found in nine of the 29 families, which are described in the article: five with A1555G, two with the T7511C, one with 7472insC and one with A3243G mutation. In the remaining 20 families, the resequencing Mitochip detected 258 mitochondrial homoplasmic variants and 107 potentially heteroplasmic variants. Controls were made by direct sequencing on selected fragments and showed a high sensibility of the MitoChip but a low specificity, especially for heteroplasmic variations. An original analysis on the basis of species conservation, frequency and phylogenetic investigation was performed to select the more probably pathogenic variants. The entire genome analysis allowed us to identify five additional families with a putatively pathogenic mitochondrial variant: T669C, C1537T, G8078A, G12236A and G15077A. These results indicate that the new MitoChip platform is a rapid and valuable tool for identification of new mtDNA mutations in deafness.
Similar content being viewed by others
Introduction
Hearing impairment is the most common sensory disorder, affecting one child in 800 to 1000 at birth. Genetic causes are estimated to represent 75% of congenital deafness. Inherited hearing impairment is a heterogenous disorder, in which about 90% of cases are non-syndromic. It can be transmitted following autosomal recessive, autosomal dominant, X-linked or maternal inheritance patterns. So far, 119 loci and 44 genes have been related to hearing impairment (Hereditary hearing Loss Homepage website).
Till now, the pathogenicity of four mitochondrial DNA (mtDNA) mutations has been clearly established in non-syndromic deafness: C1494T,1 A1555G,2 T7511C3 and A7445G.4 (MITOMAP: A Human Mitochondrial Genome Database, http://www.mitomap.org). All of these mutations are localized in the ribosomal RNA 12S gene (MTRNR1 (MIM 561000)) or in the transfer RNAser(UCN) gene (MTTS1 (MIM 590080)), and have been reported in different countries and on different haplotype backgrounds. Mutations in the 12S rRNA gene have also been related to aminoglycoside-induced deafness (MTRNR1 (MIM 580000)). Several other mutations in those genes have been related to hearing impairment but are still provisional. Mitochondrial mutations related to deafness and described on MITOMAP are reported in Table 1.
Additional factors, such as nuclear genes, environment and mitochondrial haplogroup, may modulate the phenotype. For instance, the common mtDNA mutation A1555G is strictly associated with maternally transmitted sensorineural hearing loss and represents a clear validation of the ‘two-hit’ model for nuclear–mitochondrial pathologic interaction, as the A1555G mutation is expressed in association with an additional factor such as aminoglycoside, antibiotics and nuclear modifier genes have been recently related to the disease.5, 6 Thus, the penetrance of deafness within the families can be highly variable and mitochondrial deafness is probably underestimated.
The aim of our study was to explore a cohort of deaf families living in France with a maternal inheritance pattern for mitochondrial mutations. Among 1350 families, we selected 29 families with a clear maternal pattern of inheritance and screened them for the four mtDNA mutations mentioned above. When no mitochondrial mutation could be found, we investigated the whole mitochondrial genome using a microarray resequencing chip, MitoChip version 2.0, developed by Affymetrix Inc (NASDAQ, AFFX).
Materials and methods
Families’ selection
Twenty-nine families were selected among 1350 families recruited through the national network ‘Genetic hearing impairment’ (17 centres in France) from September 2000 to December 2005 and collected in the Armand-Trousseau Children Hospital, Paris. The selection was based on genetic and phenotypic data. Families were included when deaf patients were present in at least three generations (or only two if deaf individuals were more than three per family), when maternal inheritance could be assessed and paternal inheritance excluded, and when no deafness-related mutation could be found in Gap junction Protein, Beta 2 gene (GJB2 (MIM 121011)) and Gap junction Protein, Beta 6 gene (GJB6 (MIM 604418)). Moreover, these families were selected based on the clinical phenotype with a sensorineural non-syndromic bilateral deafness, mild to profound, without any inner ear or cochlear nerve anomalies.
All patients were recruited from the genetic counseling consultation for deaf people at the Armand Trousseau Children's hospital. A complete physical examination and a personal and familial interview were performed in the proband of each family to rule out the possibility of a syndromic form of deafness and to measure the degree of hearing impairment. Audiological data were collected from the proband and his deaf-related individuals when available. Degree of hearing loss was established on the average loss at 500, 1000, 2000 and 4000 Hz as follows: normal hearing between 0 and 20 dB loss, mild hearing loss from 21 to 40 dB, moderate hearing loss from 41 to 70 dB, severe hearing loss from 71 to 90 dB and profound hearing loss more than 90 dB. One or more members of each family had an ophtalmological review (including fundoscopy), an electrocardiogram, a lacticemia, a temporal bone TDM (to exclude inner ear malformations) and tests for proteinuria and haematuria.
Blood samples were obtained from one or more affected patient in each family. Genomic DNA was extracted by standard techniques. The protocol was accepted by the Committee for the protection of individuals in Biochemical research as required by French legislation, and informed consent was obtained from all subjects or their parents.
Screening for known mtDNA mutations
mtDNA mutations C1494T, A1555G, G7444A, A7445G, T7510C, T7511C and A3243G were investigated in all patients. We investigated the A3243G mutation,7 which is usually responsible for syndromic deafness, because hearing loss can be isolated at the onset of the disease.
Those mutations were screened by DGGE and confirmed by DNA sequencing. For mutations C1494T and A1555G, PCR was performed with primers corresponding to positions 1440–1465 (forward) and 1592–1616 (reverse). For mutations in tRNA genes, PCR primers corresponded to positions 7409–7429 (forward) and positions 7529–7553 (reverse) for tRNASer(UCN) gene and positions 3163–3182 (forward) and positions 3401–3420 (reverse) for tRNALeu(UUR) gene. All forward primers had an additional GC tail at the 5′ end. Electrophoresis was performed in a 6% polyacrylamide gel with a 10–60% gradient (urea, phosphamide). Each PCR product was analysed pure and mixed with the PCR-amplified DNA of a healthy control individual corresponding to the reference sequence.
When the screening was positive with DGGE for the above three mitochondrial fragments, PCR products were subjected to direct sequencing in an ABI Prism 310 Genetic Analyser® (Applied Biosystems) to assess the presence of the mutation.
When a family was positive for A3243G mutation, urine and buccal cell were collected to determine the amount of mutation in each tissue.
For the mutations found heteroplasmic at position T7511C in tRNASer(UCN), quantification was obtained with radioactive 32P PCR-RFLP, as described by Chapiro.8
Whole-genome analysis using MitoChip 2
For selected families in which no known mitochondrial mutation was reported, a whole mitochondrial genome screening was performed with a resequencing chip: MitoChip version 2.0. (GeneChip® Mitochondrial Resequencing Array 2.0, Ref 511 300), developed by Affymetrix Inc. The MitoChip is a mtDNA resequencing chip, generated by standard photolithography and solid-phase DNA synthesis.9 About 300 000 overlapping oligonucleotide probes (25 bases), corresponding to the whole mitochondrial genome, are generated on the planed glass surface of the chip. Four oligonucleotide probes are related to every mtDNA nucleotide position, each of them differing only by the 13th base, which consists of each of the four possible nucleotides.
The subject's DNA was amplified by PCR, fragmented in 200 bp-long fragments and added to the chip for hybridization. The hybridization was revealed by fluorescence and the chip was scanned. The scanner measured the fluorescence intensity for all positions and the genotype call was generating from those raw data. The software GCOS (GeneChip Operating Software) version 1.4 can work in two conditions: haploid and diploid. In the haploid condition, only one base per genomic position is given from the raw data; in the diploid condition, two bases per genomic position can be called. The second condition would allow visualizing heteroplasmic mutations. The sequences obtained were compared with the reference mtDNA sequence, the revised Cambridge reference Sequence10 (rCRS (GenBank accession number J01415)), to define mtDNA variant. Mitochondrial haplogroups were defined on the basis of the European phylogenetic tree described by Finnilä et al.11
A double selection was applied in order to detect the most relevant variants in our deaf patients. The first was a negative selection, which excluded variants likely to be polymorphisms: the most frequent variants (more than 4% matching sequences on MTDB database,12 the D-loop variants (non-coding and highly variable region) and the conservative variants in protein-encoding genes. Then, the positive selection was applied on the basis of species conservation and literature data. Species conservation was evaluated on a compilation of 61 different mammals. The software ClustalW, available on line at http://align.genome.jp, was used for multiple sequence alignment. The criteria were less selective for the genes already implied in non-syndromic deafness than for those in which no mutations has ever been described. All the variants in the 12S rRNA gene were considered, as well as every variant in transfer RNA genes that showed more than 50% conservation. For the variants located in the 16S rRNA and protein-encoding genes, in which no mitochondrial mutation has ever been described in non-syndromic deafness, we required a conservation rate of 100% in a compilation of five mammals (Mus musculus, Rattus norvegicus, Sus scrofa, Canis familiaris and Bos taurus) and more than 60% in the compilation of the 61 mammals.
To discuss the selected mitochondrial variants, we used the following databases: Mitomap, MTDB;12 and Giib-JST mtSNP13, 14 and a phylogenetic tree of more than 2500 sequences from the Irvine's group.15
As the sensitivity and specificity of this recent molecular tool is not known, controls were performed on selected variants by conventional sequencing using the Sanger's method. The primers were designed from the website Primer3, and the PCR conditions were defined with the software ProtocolWriter on the website MutationDiscovery. Ten DNA fragments were generated to control the selected variants. Controls also targeted all variants found in the above 10 DNA fragments in haploid (36 mtDNA variants) or diploid condition (12 mtDNA variants), all no-calls (104 mtDNA positions) and no-calls succession (10), assuming that such a succession could hide an insertion or a deletion in mtDNA. A total of 162 positions were verified. Direct sequencing was performed in all maternal relatives, when available.
Results
Screening for A1555G, tRNASer(UCN) and tRNALeu(UUR) gene mutations
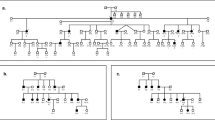
Among the 29 families, nine were found to carry a known deafness-related mtDNA mutation. These mutations were: A1555G (five families), T7511C (two families), 7472insC (one family) and A3243G (one family). The phenotypic characterization of those families is reported in Table 2 and pedigrees in Figure 1.
All the A1555G mutations were found homoplasmic on DGGE and DNA sequencing. Two deaf individuals had a history of aminoglycoside exposure. The first patient, the proband of family 1, had an accidental overdosed administration of gentamycin (9 mg/kg, three times the required dose) at 15 days of life, and bilateral profound hearing loss was diagnosed at 9 months. Her mother and grandmother did not complain of hearing loss but her two granduncles and her grandgrandmother had profound bilateral deafness, without any aminoglycoside exposure. The audiogram's shape was flat and they was no tinnitus nor vertigo associated with deafness. The second patient, a distant maternal relative of the family 2's proband, received streptomycin for a pyelonephritis. There were 25 other deaf maternal relatives in this family; as far as we know, none of them had ever received aminoglycoside. The penetrance (number of deaf maternal relatives/number of maternal relatives) was variable in the five families, ranging from 40 to 80%, with a number of deaf individuals of 4–27 per family, on two or three generations. The phenotype was also highly variable. Degree of hearing impairment ranged from mild to profound. Audiogram’ shapes were descending or flat. There was no reported history of vestibular symptoms. Onset could be congenital (two families) or deafness could occur later in life, from childhood to early adulthood (30 years). However, within a family, degree of hearing loss and age at onset were homogeneous. At last, all the probands of the five French families harbouring the A1555G mutation were originated from other countries: Israel, Spain, Iran, Portugal and Madagascar.
In the tRNASer(UCN) gene, two families were carrying the T7511C mutation and one family the 7472insC. No other reported tRNASer(UCN) gene mutation could be reported.
The two families with the T7511C mutation were previously described.8 The penetrance was 30 and 50%, respectively, but compared with family carrying the A1555G, the phenotype was far less homogeneous within the family, both in the age at onset and in the severity of the hearing impairment.
The 7472insC mutation was homoplasmic in the maternal relatives of family 8. We did not expect to find this mutation as it had only been reported in syndromic cases (hearing loss associated with neurological troubles).16, 17 The proband's deafness was diagnosed at the age of 7. The patient is now 30-year-old and has a mild hearing loss with a U-curve audiogram. His mother and a maternal cousin are deaf (respectively severe and profound hearing impairment). There was no personal or familial history of neurologic disorders.
One family of apparently non-syndromic deafness was found to carry the A3243G mutation in tRNALeu(UUR) gene. The A3243G mutation was tested in four maternal relatives of the family's proband and was always found heteroplasmic. The level of mutant was different between the tissues and between the individuals of the family (Figure 2). The severity of the disease was clearly related to the amount of mutation, as described previously.7 The proband consulted for an evolving deafness at the age of 25. The onset was in the second decade and the degree of hearing impairment was mild. It worsened quickly and the deafness became severe 5 years later. Heteroplasmic A3243G mutation was easy to find on DGGE and confirmed on DNA sequencing. Initially, deafness was thought to be sporadic, as no maternal relatives complained of hearing loss, but after audiological evaluations, the proband's mother and two aunts presented with mild bilateral hearing impairment. This mild phenotype correlated with the low level of mutant observed. There was no familial history of diabetes or neurological troubles, except migraine. Nonetheless, after the genetic diagnosis, clinical evaluation revealed the associated infraclinical symptoms of the syndromic hearing impairment related to the A3243G mutation (diabetes-deafness syndrome (MIM 520000)): glucose intolerance, slight proximal muscular weakness and hyper-lactic-pyruvic acidemia (lactic acid: 7.10 mmol/l (norm 0.5–2.2), pyruvic acid: 23.60 mg/l (norm 3–7)). Her disease evolved quickly and the muscular disorder appeared soon after the diagnosis, becoming now incapacitating.
No additional mtDNA mutation was found either in 71 additional families with maternal inheritance or in 95 sporadic cases.
Results of the whole mitochondrial genome analysis in the remaining 20 families
Twenty families did not show any mutation in the tested fragments and had their whole mitochondrial genome investigated using the resequencing MitoChip. Phenotypes were homogeneous within the families, for the degree of hearing impairment, the audiogram's shape (mostly flat or slightly descending) and the presence of associated symptoms (Table 4).
The MitoChip detected 258 variants in haploid condition. The call rate was good, between 97.2 and 98.59%, representing an average of 300 no-calls per patient. Ninety percent of those no-calls were located in C-rich regions. In term of variants, each fragment of the mtDNA was represented proportionally to its length, except for the D-loop fragment, which was unsurprisingly overrepresented (the D-loop is a fast evolving, hypervariable region). Eighteen out of these 258 variants (7%) had never been described in literature and mitochondrial databases. Forty-eight out of the 158 (30.1%) variants located in the protein-encoding genes were non-conservative.
The variants given by the chip in haploid condition allowed us to determine the family's haplogroups (Table 4). All of them belong to European haplogroups except for one family who belongs to the African haplogroup L3. The mean number of variants per haplogroup was increasing with the phylogenetic distance from the rCRS (haplogroup H2), which tends to corroborate the chip's results.
Fourteen possibly pathogenic variants were selected in the MitoChip's data according to the selection criteria. Two of them were in the 12S rRNA gene, four were in the tRNA genes and eight were in the protein-encoding genes.
We realized 45 sequences to test the 162 identified positions (see Materials and methods). Direct sequencing assessed 32 of the 36 mtDNA variants called by the chip in the haploid condition, which represents an estimated specificity of 89% for the MitoChip in haploid condition. The sensitivity of the Mitochip in haploid condition can be evaluated at 100% in our selected sample, as all the variants seen on direct sequencing had been detected by the MitoChip. All of them were found homoplasmic. The false-positive results corresponded to C-rich regions, in which the chip gave a no-call for the majority of the others patients, or to positions immediately adjacent to a mitochondrial base variation. This is due to difficulties in obtaining specific hybridization when the tested position is in the middle of a cytosines’ stretch or located adjacent to a mismatch. No genome variation could be found where the chip had given a no-call, whether in non C-rich regions (13 positions tested) or in C-rich regions (91 positions tested). One insertion was found in one of the 10 regions for which the MitoChip had given successions of no-calls.
In addition, the MitoChip found 320 variants in the diploid condition. Among them, 107 were exclusively detected with this software's configuration. The call rate was largely inferior to the haploid condition and led us to call into question the specificity of the MitoChip in the diploid condition. Sequencing controls did not detect any of the 12 controlled variants given by the chip in the diploid condition. In consequence, those results were not taken into consideration in the selection process.
Finally, the controls allowed us to confirm 10 out of the 14 selected variants. Those variants are characterized in Table 3. The phenotypes, haplogroups and pedigrees of the families in which they were described are reported in Table 4 and Figure 3.
Those 10 variants were then subjected to a more detailed analysis to determine a putative pathogenicity. As organ biopsy cannot be available, epidemiological and conservational data are important parameters to identify an mtDNA variant responsible for non-syndromic deafness. The epidemiological analysis must also include phylogenetic data, which are now available for a large range of populations, to help discriminating pathogenic mtDNA mutations and polymorphisms.18 The criteria we chose to determine the most probably pathogenic variants were as follows: the nucleotide is largely conservated, the nucleotide is very uncommon, the variant is located in a hot spot region for deafness, the variant is not linked to a single haplogroup.
Two variants were found in the 12S rRNA gene: C1537T and T669C. Both of them are considered possibly pathogenic because they are very uncommon and localized in the 12SrRNA. The C1537T variant in the 12S rRNA gene has never been recorded whether in MTDB, in Giib-JST mtSNP or in Irvine's database, but was reported in an article by Estivill et al,19 in association with the A1555G mutation in a deaf family. The T669C is localized at the opposite site of the 12S rRNA gene and has a 59% conservation rate. In MTDB database, its frequency is very low (one sequence in 2469) and we could not find this variant in a series of 130 normal hearing patients. In the Irvine's database, the T669C variant was found one time, in association with the subhaplogroup N1a, which is also our patient's haplogroup. Nevertheless, the number of patient is too minimal to rule out the pathogenicity of this mtDNA variant.
Among the two variants selected in transfer RNA genes, T7581C in tRNAAsp gene (MTTD (MIM 590015)) and G12236A in tRNASer(AGY) gene (MTTS2 (MIM 590085)), only the tRNASer(AGY) variant G12236A can be considered deleterious because T7581C is haplogroup U specific. The G12236A variant is mostly found in the African haplogroup L2, whereas our family belongs to the European haplogroup H2. It corresponds to a mismatch in the anticodon stem of the tRNASer(AGY) and has a conservational rate of 73%.
A total of six variants were located in the protein-encoding genes: three in NADH deshydrogenase 1 gene (MTND1 (MIM 516000), one cytochrome oxydase II gene (MTCO2 (MIM 516040)), one in ATPase 6 gene (MTATP6 (MIM 516060)) and one in cytochrome b gene (MTCYB (MIM 516020)). ND1 variants did not achieve to prove any pathogenicity: A3397G and A3796G have been related to Alzheimer and Parkinson diseases but are considered polymorphisms based on a scoring approach,20 C3388A is haplogroup H2 specific. G8078A is a non-conservative variant located in the cytochrome oxydase II gene. It is reported for the first time in this article. The conservation rate in a compilation of 61 mammals is 85%. It was reported in haplogroups M7 and N9, whereas our patient belongs to haplogroup U. The G8078A variant can subsequently be candidate for further investigation. Even if disrupting an initiating methionine, The A8527G in ATPase 6 gene did not show any pathogenicity on previous functional studies,21 and is haplogroup L3 specific. Thus, it is more likely a polymorphism. The G15077A variant was found in the cytochrome b gene. It had never been reported in the literature before, is uncommon in mitochondrial databases and is very well conservated (95%). It was reported in haplogroup C and P, which are not our patient's haplogroup. The G15077A is then a possibly pathogenic variant.
Finally, five were considered probably deleterious (T669C, C1537T, G8078A, G12236A and G15077A).
Discussion
Mitochondrial mutations have been implicated in non-syndromic hearing loss either as primary or as predisposing factors. Our results show that only one-third of our families with a clear maternal lineage carries one of the known mitochondrial mutations. It implies that we probably ignore many other mitochondrial deleterious variants. As 12S rRNA gene, tRNASer(UCN) gene and tRNALeu(UUR) gene are usually the only regions investigated, other mtDNA variations are simply disregarded.
The mtDNA resequencing chip MitoChip version 2.0 is a recent molecular tool proposed by Affymetrix Inc. It has been developed and used in the John Hopkins Hospital by Maitra et al.9 The first version was applied for the early detection of cancers, as specific mtDNA mutations can be regarded as tumoral markers. It was evaluated by Saladino et al22 for the mtDNA mutation detection in Leber's hereditary optic neuropathy but failed to satisfy the authors. Indeed, this version did not provide a complete sequence analysis and had an insufficient base call rate of 71–74%. It was improved in a second version, which we used, providing the sequencing of 16 544 out of the 16 569 mtDNA base pair. The call rate was also better with an average of 98% in haploid condition. The methods present two main advantages: it is time saving (48 h are required to get the sequences) and very sensitive. Zhou et al23 recently reported sequential dilutions studies, which showed that the resequencing MitoChip is able to detect as low as 2% of heteroplasmy, which is fivefold better than conventional methods. The major problem is the lack of specificity observed with the diploid condition because of numerous miscalls due to hybridization artefacts in C-rich and mismatch-adjacent regions. As there is no other method sensitive enough to authenticate the presence of very low-level heteroplasmic mtDNA variants, hybridization conditions have to be further optimized for a more extensively use of the MitoChip.
The resequencing MitoChip allowed us to investigate the whole mitochondrial genome and to identify possibly pathogenic mitochondrial variants. Determining whether or not a variant is pathogenic is a complicated process. In mitochondrial non-syndromic hearing loss, the inner ear is the only organ suffering from an energetic defect. The exact mechanism of this specific targeting is not entirely known but could be due to cochlear-specific isoforms of proteins implied in the RNA processing, which render them more sensitive to abnormal RNAs.24 As inner ear biopsies for functional studies are out of the question, the analysis of mtDNA variants relies on conservational and phylogenetical criteria. This analysis led us to find five putative pathogenic variants: T669C, C1537T, G8078A, G12236A and G15077A. Familial history of patients carrying those new variants indicates a clear maternal lineage. Furthermore, phenotypes are mostly homogeneous within the family (Table 4), which supports the hypothesis of a true mitochondrial inheritance of the disease possibly due to those variants.
Finally, our study demonstrates that the MitoChip is a reliable tool for exploring the whole mitochondrial genome and detecting new deafness-associated homoplasmic mutations.
Accession codes
References
Zhao H, Li R, Wang Q et al: Maternally inherited aminoglycoside-induced and nonsyndromic deafness is associated with the novel C1494T mutation in the mitochondrial 12S rRNA gene in a large Chinese family. Am J Hum Genet 2004; 74: 139–152.
Prezant TR, Agapian JV, Bohlman MC et al: Mitochondrial ribosomal RNA mutation associated with both antibiotic-induced and non-syndromic deafness. Nat Genet 1993; 4: 289–294.
Sue CM, Tanji K, Hadjigeorgiou G et al: Maternally inherited hearing loss in a large kindred with a novel T7511C mutation in the mitochondrial DNA tRNA(Ser(UCN)) gene. Neurology 1999; 52: 1905–1908.
Vernham GA, Reid FM, Rundle PA, Jacobs HT : Bilateral sensorineural hearing loss in members of a maternal lineage with mitochondrial point mutation. Clin Otolaryngol Allied Sci 1994; 19: 314–319.
Guan MX, Yan Q, Li X et al: Mutation in TRMU related to transfer RNA modification modulates the phenotypic expression of the deafness-associated mitochondrial 12S ribosomal RNA mutations. Am J Hum Genet 2006; 79: 291–302.
Bykhovskaya Y, Mengesha E, Wang D et al: Phenotype of non-syndromic deafness associated with the mitochondrial A1555G mutation is modulated by mitochondrial RNA modifying enzymes MTO1 and GTPBP3. Mol Genet Metab 2004; 83: 199–206.
de Vries D, de Wijs I, Ruitenbeek W et al: Extreme variability of clinical symptoms among sibs in a MELAS family correlated with heteroplasmy for the mitochondrial A3243G mutation. J Neurol Sci 1994; 124: 77–82.
Chapiro E, Feldmann D, Denoyelle F et al: Two large French pedigrees with non syndromic sensorineural deafness and the mitochondrial DNA T7511C mutation: evidence for a modulatory factor. Eur J Hum Genet 2002; 10: 851–856.
Maitra A, Cohen Y, Gillespie SE et al: The Human MitoChip: a high-throughput sequencing microarray for mitochondrial mutation detection. Genome Res 2004; 14: 812–819.
Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N : Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet 1999; 23: 147.
Finnila S, Lehtonen MS, Majamaa K : Phylogenetic network for European mtDNA. Am J Hum Genet 2001; 68: 1475–1484.
Ingman M, Gyllensten U : mtDB: Human Mitochondrial Genome Database, a resource for population genetics and medical sciences. Nucleic Acids Res 2006; 34: D749–D751.
Tanaka M, Takeyasu T, Fuku N, Li-Jun G, Kurata M : Mitochondrial genome single nucleotide polymorphisms and their phenotypes in the Japanese. Ann N Y Acad Sci 2004; 1011: 7–20.
Tanaka M, Cabrera VM, Gonzalez AM et al: Mitochondrial genome variation in eastern Asia and the peopling of Japan. Genome Res 2004; 14: 1832–1850.
Ruiz-Pesini E, Lott MT, Procaccio V et al: An enhanced MITOMAP with a global mtDNA mutational phylogeny. Nucleic Acids Res 2007; 35: D823–D828.
Cardaioli E, Da Pozzo P, Cerase A et al: Rapidly progressive neurodegeneration in a case with the 7472insC mutation and the A7472C polymorphism in the mtDNA tRNA ser(UCN) gene. Neuromuscul Disord 2006; 16: 26–31.
Hutchin TP, Navarro-Coy NC, Van Camp G et al: Multiple origins of the mtDNA 7472insC mutation associated with hearing loss and neurological dysfunction. Eur J Hum Genet 2001; 9: 385–387.
Yao YG, Salas A, Bravi CM, Bandelt HJ : A reappraisal of complete mtDNA variation in East Asian families with hearing impairment. Hum Genet 2006; 119: 505–515.
Estivill X, Govea N, Barcelo E et al: Familial progressive sensorineural deafness is mainly due to the mtDNA A1555G mutation and is enhanced by treatment of aminoglycosides. Am J Hum Genet 1998; 62: 27–35.
Mitchell AL, Elson JL, Howell N, Taylor RW, Turnbull DM : Sequence variation in mitochondrial complex I genes: mutation or polymorphism? J Med Genet 2006; 43: 175–179.
Dubot A, Godinot C, Dumur V et al: GUG is an efficient initiation codon to translate the human mitochondrial ATP6 gene. Biochem Biophys Res Commun 2004; 313: 687–693.
Saladino F, Virgilio R, Cifola I et al: Mitochondrial DNA microarray resequencing in Leber's hereditary optic neuropathy and other mitochondrial encephalomyopathies. Paper presented at: 16th Meeting of the European Neurological Society 2006, Lausanne.
Zhou S, Kassauei K, Cutler DJ et al: An oligonucleotide microarray for high-throughput sequencing of the mitochondrial genome. J Mol Diagn 2006; 8: 476–482.
Fischel-Ghodsian N, Kopke RD, Ge X : Mitochondrial dysfunction in hearing loss. Mitochondrion 2004; 4: 675–694.
Acknowledgements
This study was supported by the French Mitochondrial Disease Network, the ‘GIS-maladies rares’ and a grant provided by the French Health Department. We acknowledge the expert technical assistance of Catherine Meunier, France Michel, Corinne Chauve, Isabelle Sargis, Magalie Niasme and Marie-Ange Selva.
Author information
Authors and Affiliations
Corresponding author
Additional information
Electronic-Database Information:
Accession numbers and URLs for data presented herein are as follows:
GenBank, http://www.ncbi.nlm.nih.gov/GenBank/ for sequence information for the rCRS [accession number J01415]
OMIM: Online Mendelian Inherited in Man,
http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=OMIM, for Mitochondrial non-syndromic hearing impairment related to 12S rRNA and tRNASer(UCN) gene mutation, to aminoglycoside and for diabetes-deafness syndrome.
MITOMAP: A Human Mitochondrial Genome Database, http://www.mitomap.org
Hereditary Hearing Loss homepage; http://webhost.ua.ac.be/hhh/
Primer3, http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi
MutationDiscovery, http://www.mutationdiscovery.com
ClustalW, http://align.genome.jp/
Rights and permissions
About this article
Cite this article
Lévêque, M., Marlin, S., Jonard, L. et al. Whole mitochondrial genome screening in maternally inherited non-syndromic hearing impairment using a microarray resequencing mitochondrial DNA chip. Eur J Hum Genet 15, 1145–1155 (2007). https://doi.org/10.1038/sj.ejhg.5201891
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5201891
Keywords
This article is cited by
-
Mitochondrial genome study in blood of maternally inherited ALS cases
Human Genomics (2023)
-
Mitochondrial mutations in maternally inherited hearing loss
BMC Medical Genetics (2017)
-
Analysis of mitochondrial alterations in Brazilian patients with sensorineural hearing loss using MALDI-TOF mass spectrometry
BMC Medical Genetics (2016)
-
Frequency of mitochondrial mutations in non-syndromic hearing loss as well as possibly responsible variants found by whole mitochondrial genome screening
Journal of Human Genetics (2014)
-
Compound mitochondrial DNA mutations in a neurological patient with ataxia, myoclonus and deafness
Journal of Genetics (2014)