Abstract
Cornelia de Lange syndrome (CdLS; OMIM 122470) is a rare multiple congenital anomaly/mental retardation syndrome characterized by distinctive dysmorphic facial features, severe growth and developmental delay and abnormalities of the upper limbs. About 50% of CdLS patients have been found to have heterozygous mutations in the NIPBL gene and a few cases were recently found to be caused by mutations in the X-linked SMC1L1 gene. We performed a mutation screening of all NIPBL coding exons by direct sequencing in 11 patients (nine sporadic and two familial cases) diagnosed with CdLS in Sweden and detected mutations in seven of the cases. All were de novo, and six of the mutations have not been previously described. Four patients without identifiable NIPBL mutations were subsequently subjected to multiplex ligation-dependent probe amplification analysis to exclude whole exon deletions/duplications of NIPBL. In addition, mutation analysis of the 5′ untranslated region (5′ UTR) of NIPBL was performed. Tiling resolution array comparative genomic hybridization analysis was carried out on these four patients to detect cryptic chromosome imbalances and in addition the boys were screened for SMC1L1 mutations. We found a de novo 9p duplication with a size of 0.6 Mb in one of the patients with a CdLS-like phenotype but no mutations were detected in SMC1L1. So far, two genes (NIPBL and SMC1L1) have been identified causing CdLS or CdLS-like phenotypes. However, in a considerable proportion of individuals demonstrating the CdLS phenotype, mutations in any of these two genes are not found and other potential loci harboring additional CdLS-causing genes should be considered.
Similar content being viewed by others
Introduction
Cornelia de Lange syndrome (CdLS OMIM 122470) is a multiple congenital anomaly/mental retardation syndrome characterized by severe mental and growth retardation, distinctive dysmorphic facial features, microcephaly, hirsutism, malformations of the upper limbs, cardiac defects and gastrointestinal malformations. CdLS has a variable phenotype, including a classical and a mild type, which also evolve with age.1 The majority of the cases are sporadic, but a few cases showing an autosomal-dominant inheritance have been reported.2
Two recent studies reported mutations in the NIPBL gene to cause CdLS3, 4 and NIPBL mutations have previously been identified in 26–56% of CdLS cases.4, 5, 6, 7, 8
NIPBL is located on chromosome 5p13, consists of 47 exons and encodes delangin, a 2804 amino-acid protein which is the homolog of fungal Scc2-type sister chromatid cohesion protein and the Drosophila Nipped-B developmental regulator. NIPBL may play a role in sister chromatid cohesion in humans as precocious sister chromatid separation was identified in CdLS patients.9 The remaining cases without detectable NIPBL mutations may be explained by not yet detected mutations in NIPBL or other yet unidentified genes. An X-linked form of CdLS was recently reported in three male members from the same family and in one sporadic case, demonstrating the common combination of symptoms in the spectrum of CdLS, caused by mutations in the SMC1L1 gene which encodes a subunit of the cohesion complex.10 In addition, a large number of reports have described chromosomal abnormalities associated with CdLS, involving most chromosomes except for chromosomes 6, 15, 16, 19, 20 and 22.11
We performed a NIPBL-mutation screening by direct sequencing in a group consisting of 11 patients (nine sporadic and two familial cases) demonstrating the CdLS phenotype, in order to investigate how frequent NIPBL mutations are in the CdLS patients diagnosed in Sweden. Patients with no detectable mutation by sequencing were further investigated using multiplex ligation-dependent probe amplification (MLPA) in order to screen for whole exon deletion. In addition, the 5′ UTR of NIPBL was screened for mutations, but no abnormalities were found. All patients lacking NIPBL mutations were further investigated by tiling resolution array-based comparative genomic hybridization (array-CGH) for detection of cryptic chromosome imbalances and SMC1L1 mutation screening was performed in the boys.
Clinical evaluation
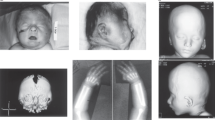
A series of subjects consisting of eleven patients diagnosed with CdLS, including nine sporadic and one familial case consisting of a brother and a sister, were included in this study after obtaining written informed consent. All patients had been referred to one of the clinical genetics departments in Sweden and diagnosed by experienced Swedish pediatricians or clinical geneticists. The clinical features of each case are summarized in Table 1 and photographs of case 1, 2, 3, 6, 10a and 10b are displayed in Figure 1. All cases were of Swedish ancestry except for cases 5 and 6, who originated from Rumania and Turkey, respectively.
Photograph of the investigated patients showing characteristic CdLS facial features. (a) case 1, (b) case 2, (c) case 3, (d) case 6, (e) case 10a and (f) case 10b. Their clinical findings are summarized in Table 1.
Methods
DNA and RNA isolation
Genomic DNA was isolated from peripheral blood lymphocytes, skin fibroblasts or EBV transformed lymphoblastoid cell lines using standard fenol extraction or Puregene blood kit (Gentra Systems Inc., Minneapolis, MN, USA) according to the manufacturer's protocol. RNA was isolated from EBV transformed lymphoblastoid cell lines using the RNAeasy kit (Qiagen,West Sussex, UK), including DNA digestion according to the manufacturer's protocol.
NIPBL and SMC1L1 mutation analysis
The entire NIPBL gene including the first noncoding exon was screened for mutations in all patients by PCR amplifications of exons 1–47 followed by bidirectional direct sequencing. PCR amplification was performed using primers previously reported by Krantz et al3 except for amplification of exons 6, 4, 33, 37 and 41 which were amplified using primers described by Miyake et al7 and exons 1, 30 and 46, for which new primers were designed (primer sequence and PCR condition available on request).
Mutation analysis of SMC1L1 was carried out on two boys without detectable NIPBL mutation by PCR amplification of exons 1–25 followed by bidirectional direct sequencing using primers previously reported by Musio et al,10 except for amplification of exons 4, 12, 19, 23 and 24, for which new primers were designed (PCR condition and primer sequence available on request). Sequence analysis was performed using ABI SeqScape software version 2.5 (Applied Biosystems).
RT-PCR
RT-PCR was carried out for cases 2 and 5 in order to detect possible disruption of the splice sites. The first strand reaction was performed from 1 μg of total RNA, using the first strand synthesis kit (Amersham). Primers for NIPBL cDNA amplification were designated in exons flanking the mutation in exon 19 or 36 in order to amplify the wild-type and the mutated allele. The PCR reaction was performed using 1 μl of cDNA using DyNAzymeEXT DNA polymerase (In vitro Sweden AB, Stockholm, Sweden) and 1.25 U products were size separated on an agarose gel along with a normal control sample in order to detect aberrant bands (primer sequence and cycling conditions available upon request).
Array-CGH
Tiling resolution array-CGH was performed on four patients (cases 8–10, Table 1) without detectable NIPBL mutations in order to detect cryptic chromosome imbalances. Microarrays with complete genome coverage containing 33 370 BAC clones were produced by the Swegene DNA Microarray Resource Center, Department of Oncology, Lund University, Sweden (http://swegene.onk.lu.se; Swegene Center Home page). The clones set consisted of the 32K BAC clone library (CHORI BACPAC Resources, http://bacpac.chori.org/genomicRearrays.php),12 additional clones located in the telomeric regions13 and clones covering microdeletion syndromes14 and printed as described previously.15 Sample labeling, hybridization and array data analysis was performed according to protocols described elsewhere.16 The detected chromosome imbalances that were not yet listed in the database of genomic variants http://projects.tcag.ca/variation/ were tested for hereditability by investigating parental samples using the same array.
MLPA
In four patients (cases 8, 9 10a and 10b), no NIPBL mutations were detected by direct sequencing. These patients were analyzed by MLPA, using kit P141 and P142, for detection of NIPBL whole exon deletions or duplications and performed according to the manufacturer's protocol (MRC Holland, Amsterdam, The Netherlands). No deletions or duplications were found in any of the patients.
For confirmation of array results, the MLPA Salsa human telomere kits P019, P020 and P036 (MRC Holland) were used and performed according to the manufacturer's protocol. Detection and size determination of the PCR fragments were carried out on a 3100-Avant™ Genetic Analyzer (Applied Biosystems).
Results
NIPBL mutations
Seven heterozygous mutations were identified in our series of 10 cases by direct sequencing of the 47 exons of the NIPBL gene. The mutations comprised of three nonsense mutations, two missense mutations, one splice mutation and one small deletion and are summarized in Table 1. All mutations were private and novel except for a nonsense mutation in exon 10 (R832X), which was previously reported in one case.6 The two missense mutations (T2146P and A2436T) altered residues that are highly conserved across species (including rat, mouse, zebra fish, Drosophila and Caenorhabditis elegans) and were not detected in 150 control subjects. In case 2, a splice site mutation was detected (IVS19+2 T>A) and in case 5 an in-frame deletion of 6 bp (GGTAGT) was detected at the 5′ end of exon 36. RT-PCR was performed in both cases in order to investigate disruption of splice sites. The results from the RT-PCR experiment are shown in Figure 2. Case 2 showed a normal band sized 335 bp, but in addition an aberrant band with a size of 254 bp. This demonstrates that the splice site mutation in case 2 results in skipping of exon 19 (size 81 bp), whereas the splicing was unaffected in case 5 as only one normal band with a size of 403 bp was detected. By sequencing the 403 bp product an in-frame 6 bp deletion was confirmed, indicating that the mutation resulted in a deletion of two amino acids (2 × valine). These residues are highly conserved across species (including rat, mouse, zebra fish, Drosophila and C. elegans).
Picture of agaose gel electrophoresis of RT-PCR product from cases 2 and 5. An aberrant band is shown in case 2 with a size of 254 bp (normal band should be 335 bp). This indicates that exon 19 (81 bp) has been skipped during splicing. Case 5 shows normal spicing of exon 36. WT=wild-type, NC=negative control.
In addition, we detected three sequence polymorphisms within the coding sequence; two previously reported in exons 10 and 33, respectively (c2021A>G;pAsn674Ser and c5874C>T;pSer1958Ser) and one novel polymorphism in exon 10 (c2772C>T;pAsn924Asn).17
Further analysis on patients without NIPBL mutations
The remaining four patients (cases 8, 9, 10a and 10b) without identifiable NIPBL mutations by direct sequencing were screened for whole exon deletions or duplications by MLPA, but no abnormalities were detected (data not shown). Mutation screening of SMC1L1 did not reveal any mutations in the two boys without NIPBL mutations.
All cases had been karyotyped by conventional G-band analysis without finding any chromosome rearrangements. Subsequently, we screened the four patients without NIPBL mutation by array-CGH using a tiling resolution BAC array for detection of cryptic chromosome abnormalities that escape detection with conventional karyotyping. With this method, we detected in addition to small genomic losses and gains that also were present in the healthy parents, a small de novo 9p24.3 duplication in case 8. The 9pter duplication was confirmed by MLPA using the commercially available 9p telomeric probes (data not shown) and had a size of 0.6 Mb, starting from clone RP11-770G15 to clone RP11-113D07 (Figure 3). Both parents showed normal chromosome 9 with array-CGH as well as MLPA (data not shown).
Array-CGH result of case 8. (a) CGH plot representing the log2 ratio of all clones located on chromosome 9 displaying a 0.6 Mb duplication on 9p24.3 consisting of six BAC clones. (b) Displays the genomic location of the 9p24.3 duplication in case 8 together with the BAC clones, genes, normal variants and other reported cases with MR in that region. *Patients reported with a duplication partly overlapping with case 8 and demonstrating phenotype including mental retardation. **Normal variants listed in database of genomic variants.
No chromosome imbalances were detected in cases 9, 10a and 10b except for losses and gains that have previously been found in healthy individuals.18 These copy number changes were considered not significant for the clinical phenotypes of our patients.
Cases 1–6 showed the classical phenotype of CdLS with severe developmental delay, limb reduction and characteristic facial features. Cases 7–9 demonstrated a milder phenotype with overlapping features of CdLS (Table 1). Only one mutation was detected in the three milder cases, a missense mutation in exon 43 in case 7 (Table 1). Case 10 was a familial case consisting of a brother and sister (cases 10a and 10b, Table 1). Both siblings were screened for NIPBL mutations by direct sequencing as well as analyzed by array-CGH to detect cryptic chromosome abnormalities. Apart from already reported polymorphisms by de Vries et al,18 no genomic imbalances were detected.
Discussion
We detected NIPBL mutations in the majority (70%) of our investigated patients, although previous mutation screening studies reported a lower detection rate (∼50%).5, 6, 7, 8 Our higher detection rate is probably due to patient selection as well as our limited sample size. Six of the patients that had a mutation in the NIPBL gene demonstrated the classical CdLS phenotype including characteristic facial features, severe growth and mental retardation and four of these cases had limb reduction (Table 1). One patient with a missense mutation had a milder phenotype with mild mental retardation, growth retardation, distinctive facial features but no limb deficiencies. Two of the patients that had no detectable NIPBL mutation (cases 8 and 9) showed some features overlapping with the CdLS phenotype (such as limb reduction and characteristic facial features) but demonstrated a clearly milder growth and mental delay compared to classical CdLS patients. Cases 10a and 10b, however, demonstrated the classical CdLS phenotypic features except for limb reduction, but no NIPBL mutation nor SMC1L1 was detected in these two siblings.
A previous report on phenotype–genotype correlation suggested that missense mutations showed a trend toward a milder phenotype compared to other types of mutations and genotype–phenotype correlation in mutation-positive and mutation-negative individuals was observed.6 However, a clear correlation between genotype and phenotype could not be confirmed in the study performed by Bhuiyan et al.8 In our study, we were not able to confirm a clear genotype–phenotype correlation. We found a missense mutation in a patient with a severe phenotype (case 6) but also in a patient with a mild phenotype (case 7) and two patients demonstrated a severe CdLS phenotype, although no NIPBL mutation was detected (cases 10a and 10b). However, the limited sample size of our study makes it difficult to perform genotype–phenotype correlation.
As chromosome abnormalities are a frequent finding in CdLS patients, we investigated the patients without detectable NIPBL mutations using tiling resolution array-CGH.
We detected a de novo small interstitial duplication on chromosome 9p24.3 in one patient, who did not demonstrate the classical CdLS phenotype but still had overlapping features with this disorder (case 8, Table 1). The size of the detected duplication is 0.6 Mb and it contains three known genes (CBWD1, DOCK8 and ANKRD15) and one predicted protein (according to Ensemble v. 36 (Feb 2006) at http://www.ensemble.org). DOCK8 and ANKRD15 have been repeatedly reported to be duplicated in healthy individuals (see database of genomic variants at http://projects.tcag.ca/variation/.) and the function of CBWD1 is not clear but might be involved in the sexual differentiation process.19 None of these genes are strong candidates to explain our patient's phenotype and it is not clear if this duplication is contributing to our patient's disorder. Several normal variants located in the duplicated region are listed in the database of genomic variants. On the other hand, two mentally retarded patients with smaller duplications located in the same region as our patients have been reported. Case 1 is a patient with mental retardation, developmental delay and infantile spasm and is listed in the Decipher database at http://www.sanger.ac.uk/PostGenomics/decipher/. Case 2 is a patient with moderate mental retardation and facial dysmorphism, which was reported by de Vries et al18 (Figure 3). From the reported data, it is not clear if the duplications were inherited from the parents, neither is it clear if the duplications were considered to be associated with the patient's phenotype.
Recent developments in genome-scanning technologies and computational methodologies, and the availability of a reference sequence for comparison, have made it possible to detect structural variants in the human genome with or without clinical symptoms. At this point, we cannot determine if the duplication found in our patient is a clinical significant chromosome abnormality or a normal genomic variant.
In general, in a considerable number of patients demonstrating the CdLS or CdLS overlapping phenotype no NIPBL mutations can be detected. It might be possible that NIPBL mutations are not detected in a large proportion of the CdLS cases because mutation detection is complicated by the large size of the gene. The methods used so far for mutation screening of CdLS patients, like DHPLC or CSGE followed by direct sequencing (alternatively only direct sequencing) of the coding regions including intron–exon boundaries, have not ruled out whole exon deletions or mutations in regulatory elements. Large deletions involving the whole gene have been investigated by FISH or array-CGH and have so far not been detected.5, 7 In our study, we performed a comprehensive mutation screening of NIPBL by screened for mutations in all exons including the nontranslated first exon as well as investigated whole exon deletions or duplications using MLPA in all patients negative for NIPBL mutations. To exclude larger deletions array-CGH containing BAC clones (RP11-169M01 and RP11-715N02 ) covering NIPBL was used. No deletions or other rearrangements were found in the NIPBL region.
The recent report on X-linked CdLS caused by SMC1L1 mutations, motivated us to investigate the boys in our study where despite our extensive laboratory investigation no etiology was found for their disorder. However, no SMC1L1 mutations were identified in the two investigated cases in this study.
In conclusion, NIPBL mutations are detected in the majority of individuals demonstrating the classical CdLS phenotype and chromosome abnormalities found in CdLS individuals may result in phenocopies or might be a coincidental finding and unrelated to the observed phenotype. However, the recent identification of SMC1L1 mutations in patients diagnosed with CdLS, implies that locus heterogeneity is present for CdLS.
References
Allanson JE, Hennekam RC, Ireland M : De Lange syndrome: subjective and objective comparison of the classical and mild phenotypes. J Med Genet 1997; 34: 645–650.
Russell KL, Ming JE, Patel K, Jukofsky L, Magnusson M, Krantz ID : Dominant paternal transmission of Cornelia de Lange syndrome: a new case and review of 25 previously reported familial recurrences. Am J Med Genet 2001; 104: 267–276.
Krantz ID, McCallum J, DeScipio C et al: Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat Genet 2004; 36: 631–635.
Tonkin ET, Wang TJ, Lisgo S, Bamshad MJ, Strachan T : NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat Genet 2004; 36: 636–641.
Borck G, Redon R, Sanlaville D et al: NIPBL mutations and genetic heterogeneity in Cornelia de Lange syndrome. J Med Genet 2004; 41: e128.
Gillis LA, McCallum J, Kaur M et al: NIPBL mutational analysis in 120 individuals with Cornelia de Lange syndrome and evaluation of genotype-phenotype correlations. Am J Hum Genet 2004; 75: 610–623.
Miyake N, Visser R, Kinoshita A et al: Four novel NIPBL mutations in Japanese patients with Cornelia de Lange syndrome. Am J Med Genet A 2005; 135: 103–105.
Bhuiyan Z, Klein M, Hammond P et al: Genotype–phenotype correlations of 39 patients with Cornelia de Lange syndrome: the Dutch experience. J Med Genet 2006; 43: 568–575.
Kaur M, DeScipio C, McCallum J et al: Precocious sister chromatid separation (PSCS) in Cornelia de Lange syndrome. Am J Med Genet A 2005; 138: 27–31.
Musio A, Selicorni A, Focarelli ML et al: X-linked Cornelia de Lange syndrome owing to SMC1L1 mutations. Nat Genet 2006; 38: 528–530.
DeScipio C, Kaur M, Yaeger D et al: Chromosome rearrangements in Cornelia de Lange syndrome (CdLS): report of a der(3)t(3;12)(p25.3;p13.3) in two half sibs with features of CdLS and review of reported CdLS cases with chromosome rearrangements. Am J Med Genet A 2005; 137: 276–282.
Krzywinski M, Bosdet I, Smailus D et al: A set of BAC clones spanning the human genome. Nucleic Acids Res 2004; 32: 3651–3660.
Knight SJ, Lese CM, Precht KS et al: An optimized set of human telomere clones for studying telomere integrity and architecture. Am J Hum Genet 2000; 67: 320–332.
Vissers LE, de Vries BB, Osoegawa K et al: Array-based comparative genomic hybridization for the genomewide detection of submicroscopic chromosomal abnormalities. Am J Hum Genet 2003; 73: 1261–1270.
Jonsson G, Naylor TL, Vallon-Christersson J et al: Distinct genomic profiles in hereditary breast tumors identified by array-based comparative genomic hybridization. Cancer Res 2005; 65: 7612–7621.
Schoumans J, Staaf J, Jonsson G et al: Detection and delineation of an unusual 17p11.2 deletion by array-CGH and refinement of the Smith–Magenis syndrome minimum deletion to approximately 650 kb. Eur J Med Genet 2005; 48: 290–300.
den Dunnen JT, Antonarakis SE : Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum Mutat 2000; 15: 7–12.
de Vries BB, Pfundt R, Leisink M et al: Diagnostic genome profiling in mental retardation. Am J Hum Genet 2005; 77: 606–616.
Wade J, Tang YP, Peabody C, Tempelman RJ : Enhanced gene expression in the forebrain of hatchling and juvenile male zebra finches. J Neurobiol 2005; 64: 224–238.
Acknowledgements
We thank the patients and their parents for participating in this study. This work was supported by funds from the Swedish Research Council, Frimurarna Barnahuset Foundation, Linnea och Josef Carlsson Foundation and Kronprincessan Lovisa Foundation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Schoumans, J., Wincent, J., Barbaro, M. et al. Comprehensive mutational analysis of a cohort of Swedish Cornelia de Lange syndrome patients. Eur J Hum Genet 15, 143–149 (2007). https://doi.org/10.1038/sj.ejhg.5201737
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5201737
Keywords
This article is cited by
-
Clinical and genetic study of 20 patients from China with Cornelia de Lange syndrome
BMC Pediatrics (2018)
-
Transcriptomics analysis revealing candidate networks and genes for the body size sexual dimorphism of Chinese tongue sole (Cynoglossus semilaevis)
Functional & Integrative Genomics (2018)
-
Genomic imbalances in patients with a clinical presentation in the spectrum of Cornelia de Lange syndrome
BMC Medical Genetics (2013)
-
Spectrum of NIPBL gene mutations in Polish patients with Cornelia de Lange syndrome
Journal of Applied Genetics (2013)
-
Intragenic and large NIPBL rearrangements revealed by MLPA in Cornelia de Lange patients
European Journal of Human Genetics (2012)