Abstract
Pulmonary arterial hypertension is a progressive disease, characterised by increased proliferation of pulmonary artery smooth muscle cells, vasoconstriction and remodelling of the vascular wall leading to right heart failure and death. The idiopathic form is rare (idiopathic arterial primary hypertension (IPAH); formerly PPH, MIM# 178600). Our group correlated a deficiency in vasoactive intestinal peptide (VIP; MIM# 192320) levels in serum and lung tissue with the pathogenesis of IPAH. The aim of this study was to investigate the relevance of genetic alterations in VIP to the development of IPAH. We screened 10 patients (age 4–66 years) for alterations in the coding, the noncoding regions and the enhancer region of the VIP gene by direct sequencing. In eight of 10 patients, we found alterations compared to the wild-type sequence. We detected nine alterations. In the noncoding regions, eight alterations were in the introns 1, 2, 3 and 4 (g.448G>A g.501C>T g.764T>C g.2267A>T g.2390C>T g.3144T>C g.3912A>G g.4857A>G). In the coding regions, a single alteration in the 3′ untranslated region in exon 7 (g.8129T>C) was observed in five patients. It appeared in 46% of the control group. The frequency of this alteration in the coding region of the VIP gene could therefore not be correlated with the appearance of IPAH. Apart from the importance of VIP signalling, genetic and/or environmental modifiers might therefore contribute to the development and perpetuation of the disease.
Similar content being viewed by others
Introduction
Idiopathic arterial primary hypertension (IPAH, formerly PPH) is a rare progressive disease with unknown aetiology and a multifactorial pathophysiology.1 The incidence is one to two cases per million. Recently, we identified a low vasoactive intestinal peptide (VIP) expression in patients with IPAH evidenced by reduced VIP levels in the lung tissue and serum of IPAH patients.2 VIP is a 28 amino-acid peptide.3 The human VIP gene is a 8 837 bp single copy gene on chromosome 6q24 comprising seven exons and six introns with each coding exon producing a distinct functional domain in the protein.4 The gene codes for a high molecular weight precursor protein also containing the histidine methionine amide peptide hormone (peptide histidine methionin-27 (PHM-27)). Exon 1 encodes the 5′ untranslated region (5′UTR) of the prepro-VIP/PHM-27 mRNA, exon 2 the signal peptide and exon 3 the amino-terminal region of the pro-VIP/PHM-27. Exon 4 encodes PHM-27, exon 5 VIP, exon 6 the carboxyterminal region of the precursor and exon 7 the 3′ untranslated region of the mRNA.5
Several considerations gave rise to this study: VIP was lowered in serum and lung tissue of IPAH patients as demonstrated by Petkov et al.2 In addition, there exists also a familial form of the disease. As VIP knockout mice show alterations in the pulmonary circulation reminiscent of IPAH (personal communication, Sami Said, 2005) we sequenced the entire gene in IPAH patients.
Methods
Individuals
IPAH patients (n=10, Caucasian) were recruited with characteristics summarised in Table 1. Diagnosis was established according to the NYHA classification of the National Institutes of Health Registry.6 For serum VIP determination blood was obtained by venipuncture in a closed vacuum system containing trasylol as protease inhibitor (500 U/ml blood). VIP concentrations were determined by means of a commercially available radioimmunoassay (RIA; DiaSorin, Stillwater, MN, USA). Detection thresholds were at either 5 or 10 pg/ml. The latter was used in previous work.2
Randomised control samples (n=50) were obtained from the same population. Informed consent was obtained from all participating individuals (patients and controls) or their representatives and the study protocol was approved by the Austrian ethical review board.
Screening for mutations in the VIP gene by PCR and sequencing
Genomic DNA was extracted from peripheral blood with an ‘EZ NA Blood DNA Kit II’ kit (Peqlab, Erlangen, Germany). The VIP gene was amplified in a GenAmp thermocycler (Applied Biosystems, Perkin Elmer, Foster City, CA, USA) using the primer sets with annealing temperatures for 30 s in brackets: C2 for 5′-TTTTCTTCTTATCATTCAGCAG-3′, rev 5′-CTCGCCCAGTCGTGCTC-3′ (58°C); C3 for 5′-AATGGCCCTGGCTATAACAC-3′, rev 5′-CAAAAGCAAATGTTCCCACTG-3′ (51°C); C4 for 5′-AGATGCTTTACCCTGCTTGC-3′, rev 5′-CCTTTTCACCTGCTGTCTGG-3′ (56°C); C5 for 5′-TCTTGAAGCTGTTTCCTTCG-3′, rev 5′-TCACCATGTTTGGGAATGTG-3′ (43°C); C6 for 5′-TCAGGGCATGAGAGACACTG-3′, rev 5′-TGATTTGGGAGTTGGGAGTC-3′ (55°C); C7 for 5′-TGGGTTGGTTTCCTTCTTTG-3′, rev 5′-CCCCAA-GCTTTGCTTCAAAT-3′ (43°C); C8 for 5′-TGACTCCCAACTCCCAAATC-3′, rev 5′-TG-CTCCCTCAAAGGGTATTC-3′ (45°C); C9 for 5′-TTGCGGAACCATACACACTG-3′, rev 5′-TGTCTGCCCCACTGAATAAAG-3′ (51°C); C10 for 5′-ATGGAAGAGTTGCCAAGGAG-3′, rev 5′-AGTCCATGGGCTTGATAGATG-3′ (48°C); C11 for 5′-TGCTGATGGAGTTTTCACCA-3′, rev 5′-CAGGTTTCTTGGAGCCACAT-3′(54°C); C12 for 5′-TTCCTT-GCTTTCCTTCATCC-3′, rev 5′-CATCTAGTGGGCAAAGAGGTC-3′ (47°C); C13 for 5′-CCGCCTTAGAAAACAAATGG-3′, rev 5′-TGCACGAATATACCCACCAC-3′ (49°C); C14 for 5′-CCCCATACACTTTCAGAGCAC-3′, rev 5′-TTTCATAGTAATGGGCCATGC-3′ (49°C); C15 for 5′-AATGGGCTTTCCTTTTGAGG-3′, rev 5′-TTCTTTGAGCAAAGGGACTTG-3′ (48°C); C16 for 5′-CTGAAGGTGAGCCAGATTCC-3′, rev 5′-GAAATTGC-AGGCTTTTTATGAG-3′ (47°C) and C17 for 5′-TGGATTGCTAATCACCAAAGG-3′, rev 5′-ACATTTCCCAGCCTCTTCTTG-3′(48°C). The enhancer sequence of 213 bp located 4.4 kb upstream from the transcription start site was amplified with the primer set C1 for 5′-GAGTTGAAGGCTGGTTGCAT-3′, rev 5′-GGTTGTATGTATCAGACGATTTCC-3′ (60°C). Extension times were 72°C for 90 s and PCR was performed for 35 cycles. The reaction mix contained 200 μ M dNTPs, 1.5 mM MgCl2, 20 mM Tris-HCl (pH 8.0), 50 mM KCl and 1 U Platinum Taq polymerase (Invitrogen, San Diego, CA, USA). PCR products were separated on 1% agarose gels containing 0.5 μg/ml ethidium bromide at 1 V/cm. After purification of the PCR products (QIAquick PCR Purification Kit, Hilden, Germany), all amplificants were sequenced on both strands with the PCR primers on an ABI-Prism 3100 Genetic Analyser (Applied Biosystems) and compared at the NCBI interface http://www.ncbi.nlm.nih.gov/blast/bl2seq/bl2.html to the wild-type sequence retrieved from the genomic GenBank chromosome 6 reference sequence contig (NT_025741.13). Alterations were numbered from the transcriptional start site of the human VIP gene (NM_003381.2). DNA from control individuals was amplified with the C17 primer set and the 3′untranslated region (3′UTR) region (exon 7) sequenced.
Statistics
The alteration g.8129T>C. was the only one to be found in a higher number of patients (see Results). Therefore, the frequency of this alteration in the coding region was compared between patients and the control group with the χ2 test. VIP serum levels of IPAH patients were compared to a previously published control sample2 by Student's T-test.7 P<0.05 was considered significant.
Results
Role of VIP serum level in IPAH
VIP serum levels of our IPAH patient group were as low as 13.16±8.5 pg/ml, when excluding patient 2. This patient was measured twice to confirm the 10-fold higher VIP-level (Table 1) as compared to normal control levels of 42.4±17.1 pg/ml2. The difference between our IPAH patients and the controls2 was significant (P<0.05).
Genetics in IPAH
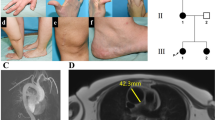
Analysis of the complete coding and non-coding region of the VIP gene of 10 patients revealed several alterations from the nucleotide sequence in comparison to the wild type (wt) sequence. Eight of 10 tested patients were carriers of different homozygous (hom) and heterozygous (het) nucleotide changes, examples of each found alteration are shown in Figure 1. Alterations were found in introns 1–4 and exon 7. In intron 1, heterozygous g.448G>A was found in patient no.7 (a) and heterozygous g.501C>T in patient no.6 (b). Moreover, a homozygous g.764T>C alteration was detected in patient no. 1 and heterozygous g.764T>C was found in patient no. 5 (c). In intron 2, we detected homozygous g.2267A>T in patient no. 1 (d). Homozygous g.2390C>T in patient no.1 and heterozygous g.2390C>T in patients 5, 6 and 8 were detected (example patients no. 1 and 6 are shown in Figure 1e). Patient no. 1 carried a homozygous g.3144T>C, whereas patient no. 5 showed a heterozygous g.3144T>C genotype (f). A heterozygous g.3912A>G alteration located in intron 3 was found in patients 6 and 8 (patient no. 8 is shown in Figure 1(g). In intron 4, homozygous g.4857A>G was found in patient no. 2 and heterozygous g.4857A>G in patient no. 7 (h).
Electrophoregrams of altered regions in the VIP gene of IPAH-paptients in comparison to the wild-type sequence. Electrophoregrams of altered regions in the VIP gene are shown in comparison to the wild-type sequence (wt). Alterations in intron 1 are het g.448G>A in no.7 (a), het g.501C>T in no.6 (b), hom (no.1) and het (no.5) g.764T>C shown in (c). Alterations in intron 2 are hom g.2267A>T (d) in no.1, hom (no.1) and het (no.6) g.2390C>T (e) and hom (no.1) and het g.3144T>C in no.5 (f). Het g.3912A>G alteration in no.8 (g) is located in intron 3 and hom (no.2) and het g.4857A>G (no.7) alterations (h) are in intron 4. Alterations in exon 7 (i) are either hom (no.1) or het g.8129T>C (no.3).
In the entire exonic region, a single g.8129T>C alteration was observed in five patients, it was homozygous in patients 1 and 8 and heterozygous in patients 3, 6 and 7 (examples: hom. and het., shown for patients 1 and 3 in Figure 1i). This alteration was further analysed by screening of 50 control DNA samples by direct PCR sequencing. In total, g.8129T>C was observed in 46% (23 individuals). Statistical analysis revealed no significant difference between the two samples.
The sequencing of the known 213 bp enhancer element containing fragment far upstream from the transcription start site did not reveal any hints for alterations in this promoter region sequence (data not shown).
Discussion
We could not verify a VIP gene alteration related to IPAH. We found lowered VIP serum concentrations, except for one exaggerated VIP level. For this one patient we speculate that it may reflect a compensatory upregulation due to a receptor defect.
The serum levels were determined by RIA, they reflect the tissue levels of the neuropeptide VIP.2 In the present study, we used also a RIA with a lower detection threshold. With this method, detection of 5 pg/ml can be measured and discrete figures instead of ‘not detectable’ values in the very low concentration range are obtained. The RIA revealed an average VIP serum level which was at 13.16±8.5 pg/ml when excluding patient no. 2 with the 10-fold increase (reference in normal controls: 42.4±17.1 pg/ml2). The average serum level was significantly (P<0,05) below the published serum level in control populations, which was 42.4±17.1 pg/ml.2 The VIP serum level of patient no. 2 was over 420 pg/ml. This extremely elevated value may indicate that there are two types of the disease, characterised by either low or high VIP serum levels. The high concentration could reflect a compensatory upregulation of the peptide concomitant with an ineffectiveVIP signalling.
Other genetic backgrounds of IPAH have been investigated in genes like bone morphogenetic protein receptor type II (BMPRII) or activins receptor-like kinase-1 (ALK-1).8 Only 10% of sporadic PAH cases have BMPRII mutations. In the hereditary form each detected mutation is unique to a family. Therefore, no correlation of particular BMPRII alterations could be associated with IPAH.9 In the search for gene alterations, we opted for direct sequencing of important parts of the VIP-promotor, of the whole coding and non-coding regions. In a pilot study, we found alterations in patients as shown in Figure 1. To our knowledge the heterozygous and homozygous alterations in introns 1, 2, 3 and 4 and exon 7 (Figure 1) of the VIP gene are novel. None of the intronic alterations occurred in known splicing consensus motifs. For this we conclude that the occasional findings are not relevant alterations.
The only alteration with a higher frequency was found in five out of 10 patients. This was the g.8129T>C alteration located in the 3′UTR region of exon 7. As the control group revealed a similar percentage (46%) when using the same methodology, we believe that this alteration cannot be related to IPAH. It seems to be a common modification of the nucleotide sequence.
A fragment in the promotor region with two enhancer elements far upstream from the transcription start site has been described.10 We found no hints for alterations in this promoter region sequence in our patient population.
VIP and PHM-27 are parts from the same precursor protein, located on adjacent exons in the human genome.11 Both peptides are structurally related with similar biological acitvity in the lung.12 Hence, we screened both genes for their role in the disease,13 none of them carried a gene alteration associated with IPAH.
VIP activity is mediated by three different G-coupled receptors VPAC1, VPAC2, and PAC1. VIP knockout mice show alterations in the pulmonary circulation reminiscent of IPAH (Sami Said, personal communication, 2005). So far no data from VPAC1 knockout mice are available. VPAC2 deficient mice do not develop IPAH.14 PAC-1 deficient mice reveal a pathology similar to IPAH, that is increased muscularisation of small pulmonary vessels and elevated right ventricular end-diastolic pressure leading to right heart failure.15 We therefore, conclude that in IPAH the VIP signalling pathway failure concerns PAC1, and/or possibly VPAC1. Apparently IPAH is a complex genetic disease. Therefore, the interaction of different genetic modifiers in combination with environmental risk factors might give more insight into the initiation of the disease.
Accession codes
References
Farber HW, Loscalzo J : Pulmonary arterial hypertension. N Engl J Med 2004; 351: 1655–1665.
Petkov V, Mosgoeller W, Ziesche R et al: Vasoactive intestinal peptide as a new drug for treatment of primary pulmonary hypertension. J Clin Invest 2003; 111: 1339–1346.
Said SI, Mutt V : Polypeptide with broad biological activity: isolation from small intestine. Science 1970; 169: 1217–1218.
Gozes I, Bodner M, Shwartz H, Shani Y, Fridkin M : Studies toward the biosynthesis of vasoactive intestinal peptide (VIP). Peptides 1984; 5: 161–166.
Yamagami T, Ohsawa K, Nishizawa M et al: Complete nucleotide sequence of human vasoactive intestinal peptide/PHM-27 gene and its inducible promoter. Ann NY Acad Sci 1988; 527: 87–102.
Rich S, Dantzker DR, Ayres SM et al: Primary pulmonary hypertension. A national prospective study. Ann Intern Med 1987; 107: 216–223.
Sokal RR, Rohlf FJ : Biometry. W.H. Freeman and Company: New York, 1981.
Loscalzo J : Genetic clues to the cause of primary pulmonary hypertension. N Engl J Med 2001; 345: 367–371.
Newman JH, Trembath RC, Morse JA et al: Genetic basis of pulmonary arterial hypertension: current understanding and future directions. J Am Coll Cardiol 2004; 43: 33S–39S.
Liu D, Krajniak K, Chun D et al: VIP gene transcription is regulated by far upstream enhancer and repressor elements. Biochem Biophys Res Commun 2001; 284: 211–218.
Itoh N, Obata K, Yanaihara N, Okamoto H : Human preprovasoactive intestinal polypeptide contains a novel PHI-27-like peptide, PHM-27. Nature 1983; 304: 547–549.
Barnes PJ : Neuropeptides in human airways: function and clinical implications. Am Rev Respir Dis 1987; 136: S77–S83.
Schulte-Bockholt A, Meier DA, Hopp K et al: Gene expression and transcript size of the prepro-peptide VIP/PHM-27 in normal human tissue. Neurosci Lett 1993; 164: 101–104.
Asnicar MA, Koster A, Heiman ML et al: Vasoactive intestinal polypeptide/pituitary adenylate cyclase-activating peptide receptor 2 deficiency in mice results in growth retardation and increased basal metabolic rate. Endocrinology 2002; 143: 3994–4006.
Otto C, Hein L, Brede M et al: Pulmonary hypertension and right heart failure in pituitary adenylate cyclase-activating polypeptide type I receptor-deficient mice. Circulation 2004; 110: 3245–3251.
Acknowledgements
We thank the patients and their relatives for cooperation. We thank D Bonderman for providing patient material and DS Habringer for excellent technical assistance. This research was supported by grants from the Jubiläumsfonds of the Austrian National Bank (Grant no. 11211) and from the European Commission contract (Grant no. QLK3-CT-2002-02031).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Haberl, I., Frei, K., Ramsebner, R. et al. Vasoactive intestinal peptide gene alterations in patients with idiopathic pulmonary arterial hypertension. Eur J Hum Genet 15, 18–22 (2007). https://doi.org/10.1038/sj.ejhg.5201711
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5201711
Keywords
This article is cited by
-
Prospect of vasoactive intestinal peptide therapy for COPD/PAH and asthma: a review
Respiratory Research (2011)