Abstract
Alport syndrome (AS) is a genetically heterogeneous renal hereditary disease. Male-to-male transmission has been considered fully indicative of autosomal dominant AS. We report a family with male-to-male transmission of X-linked AS due to an extra X chromosome of paternal origin in the proband. Linkage analysis excluded the autosomal loci and demonstrated segregation with the COL4A5 locus (Xq22.3). Sperm FISH analysis from his father detected an increased XY disomy. Mutation screening of the COL4A5 gene identified a splicing mutation, c.4688G>A. The proband and his paternal grandmother showed random X chromosome inactivation. However, a preferential expression of the aberrantly spliced transcript was detected in the proband when compared to his grandmother. This finding could explain why the AS phenotype of this 47,XXY boy resembles more an affected male than a female carrier. This is the first reported case of concurrence of Alport and Klinefelter syndromes.
Similar content being viewed by others
Introduction
Alport syndrome (AS) is a hereditary type IV collagen disease that leads to hematuria and eventually to end-stage renal disease (ESRD). Moreover, these renal alterations are often associated with sensorineural hearing loss and ocular lesions. The estimated prevalence of the disease is 1:5000.1
This disorder is genetically heterogeneous and can be transmitted following an X-linked or an autosomal pattern of inheritance. X-linked AS (MIM 301050) is the most frequent form of the disease and is caused by mutations in the COL4A5 gene at Xq22.3.2 Autosomal recessive AS (MIM 203780) and autosomal dominant AS (MIM 104200) are due to mutations in the COL4A3 and COL4A4 genes,3 both located at chromosome 2q36–q37. The existence of male-to-male transmission in an AS pedigree has been considered fully indicative of autosomal dominant AS, in which men are as affected as much as women.4 On the other hand, X-linked AS male patients have a more severe phenotype than female carriers, who have variable clinical features ranging from isolated microhematuria to renal impairment, though this occurs at an older age than in men.
Klinefelter syndrome (KS) is the most common genetic cause of human male infertility with a prevalence of one in 500–1000 males. It is genetically defined by the presence of an extra X chromosome (47,XXY) and, clinically, it is characterized by small testes, gynecomastia, hypogonadism and higher than normal concentrations of follicle-stimulating hormone.5 Approximately half of 47,XXY males arise because of a maternal meiotic error, either during the first or second division, and the other 50% are due to a nondisjunction of the paternal sex chromosomes during meiosis I.6
Here, we describe the molecular and cytogenetic characterization of a family with male-to-male transmission of X-linked AS.
Subjects and methods
Patients
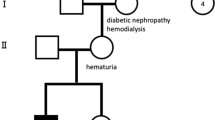
The proband, a 5-year-old boy, was referred for AS testing due to presenting with macrohematuria from 2 years of age, proteinuria of 9 mg/h/m2 and having familial antecedents of the disease. His physical examination was normal, with average genital development for his age and no signs of hypoacusia, although due to his early age no audiometric examination had been carried out. His paternal grandmother entered ESRD, after a period with proteinuria and hematuria, at 59 years of age. The father and an uncle of the proband had macrohematuria since early childhood with progressive proteinuria and entered ESRD at 21 and 17 years of age, respectively (Figure 1a). The diagnosis of AS was confirmed by finding the typical AS glomerular basement membrane ultrastructural abnormalities in a renal biopsy from the father of the proband. Informed consent was obtained from all family members participating in the study and our institutional ethics committees approved the procedures.
Linkage analysis. (a) Pedigree of the AS family showing linkage to markers flanking and within the COL4A5 gene (Xq22–q23) and nonsegregation with the COL4A3/COL4A4 (2q36–q37) loci. The haplotype associated with the disease is boxed. The proband (III-1, arrow) has two X chromosomes. (b) Fluorescent PCRs of microsatellites DXS1120 (A), DXS456 (B), DXS6797 (C) and DXS6802 (D) at Xq22-q23 demonstrating the detection of paternal (II-3) and maternal (II-4) alleles in the proband with KS (III-1). Peaks highlighted in black identify the A, B and D marker specific-alleles and those in grey correspond to marker C.
DNA/RNA extraction
Genomic DNA was obtained from peripheral blood with standard methods. Total RNA was extracted from hair roots, peripheral blood and buccal mucosa cells using the TRIzol® reagent (Invitrogen, Paisley, UK), according to the manufacturer's instructions.
Linkage analysis
Linkage analysis was performed using polymorphic markers spanning the COL4A3/COL4A4 and COL4A5 loci. Markers used for the analysis of the COL4A3/COL4A4 region were: D2S351-2.10-D2S159-0.24-D2S2158 (IVS1)-0.20-D2S401-1.48-D2S362. Microsatellites used for the COL4A5 locus were: DXS1120-0.86-DXS6797-0.27-DXS6802 (IVS1)-0.19-COL4A5-2B6 (3′UTR)-0.64-DXS1210-0.59-DXS456 (the number between markers denotes intermarker distance expressed in megabases; intragenic marker localization is indicated in brackets). Primer sequences were obtained from the GDB Human Genome Database (www.gdb.org) and one of each set of primers was fluorescently labelled. Amplification conditions were performed at annealing temperatures between 51 and 62°C using the PCR Master Mix reagent (Promega, Madison, WI, USA). PCR products were run on an ABI PRISM 3100-Avant genetic analyzer (Applied Biosystems, Foster City, CA, USA).
Reverse transcription
Total RNA from hair roots (0.5 μg), peripheral blood (2 μg) and buccal mucosa cells (1 μg) was reverse transcribed with the High-Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA, USA) using random hexamers under conditions recommended by the manufacturer.
COL4A5 mutation analysis
The entire hair root COL4A5 cDNA was amplified in 10 overlapping fragments ranging in size from 513 to 890 bp (Tazón-Vega et al, manuscript in preparation). All RT-PCR products were run on a 2% agarose gel and automatically sequenced using the Big Dye DNA Sequencing kit v2.0 (Applied Biosystems, Foster City, CA, USA) and an ABI PRISM 3100-Avant genetic analyzer. An alteration was detected in the most 3′ fragment of the COL4A5 cDNA (containing exons 47–51). This cDNA alteration was characterized at the genomic level by amplifying and sequencing exon 48 of the COL4A5 gene, using primers and conditions described by Renieri et al.7 Agarose gel electrophoresis of the abnormal cDNA fragment resulted in two distinct bands (wild-type and mutated transcripts, see Results section) in the family members bearing two X chromosomes. Semiquantitative RT-PCR analysis of both transcripts was carried out as follows. PCR amplifications were performed with a new set of primers flanking exon 48 to reduce the cDNA amplicon size (forward: 5′-AGAGCCC ACGGTCAAGACTT-3′, located in exon 47, and reverse: 5′-TGCCCCTGCACTTGTATGC-3′, located in exon 49) using the following conditions: 60 s at 95°C, 45 s at 58°C and 60 s at 72°C. PCRs were carried out for a number of cycles ranging from 20 to 40. We determined that from 26 to 30 cycles we had not reached the stationary phase of the PCR. These RT-PCR products were run on a 2% agarose gel and the two resulting bands were quantified using the Quantity One Software (Biorad, Hercules, CA, USA).
X chromosome inactivation analysis
Since only genes on the active X chromosome are expressed, we studied the pattern of X chromosome inactivation by transcriptional analysis of the polymorphic CAG repeat of the Androgen Receptor (AR) gene. First, we amplified this polymorphism using genomic DNA from the proband and his paternal grandmother to determine if they were heterozygous and to study the level of preferential amplification of the smaller alleles. We used previously described primers8 and 25 PCR cycles of 90 s at 95°C and 120 s at 65°C. Once their heterozygosity was confirmed, RNA isolated from the hair root cells of the AS-KS patient and from the hair roots, peripheral blood and buccal mucosa cells of his grandmother was studied by AR RT-PCR, using the aforementioned primers and PCR conditions except for the number of cycles which were 30. DNA and RNA amplification products were run on an ABI PRISM 3100-Avant and analysed by Genescan software for the quantification of each allele by peak area. X-inactivation was calculated as [(Ac1/Ag1)/(Ac1/Ag1+Ac2/Ag2)] × 100, where Ac1 and Ac2 refer to the peak areas of the larger and smaller cDNA alleles, respectively, and Ag1 and Ag2 are the corresponding peak areas of the genomic alleles (used to correct for preferential amplification of smaller alleles).9
Karyotype analysis
Metaphase spreads were obtained from peripheral blood lymphocytes from the proband and his parents and were analysed by G-banding.
Sperm FISH analysis
Semen samples were collected from the father of the proband (33 years old) and eight healthy control donors (33–58 years old) after obtaining informed consent from all individuals. At the moment of collection the father was on dialysis. Four-colour FISH analysis with DNA probes specific to chromosomes 6, 21, X and Y was carried out. Procedure details, probes characteristics, scoring criteria and statistical analysis were as previously described.10
Results
Linkage analysis for the COL4A3/COL4A4 loci was carried out and no segregation with the disease was identified (Figure 1a). Then, linkage to COL4A5 was tested and it was confirmed. The proband had an extra X chromosome of paternal origin giving rise to the concurrence of both Alport and Klinefelter syndromes (Figure 1a and b).
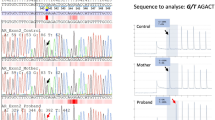
Sequencing of the whole COL4A5 coding region detected exon 48 skipping in the most 3′ fragment of the COL4A5 cDNA in all affected members of the family. Agarose gel electrophoresis of this fragment showed a preferential expression of the mutated COL4A5 transcript in the proband compared to his grandmother. Semiquantitative RT-PCR analysis to determine the relative expression levels of the wild-type and aberrantly spliced transcripts showed 50% of the correctly spliced transcript in the proband and 92% in his grandmother (Figure 2a). Characterization of the mutation at the genomic level identified the c.4688G>A substitution in the last nucleotide of exon 48 (Figure 2b). This mutation cosegregated with the disease since it was detected in all four affected family members and not in healthy individuals.
COL4A5 mutation characterization. (a) Semiquantitative RT-PCR analysis of wild-type (wt, 333 bp) and aberrantly spliced (155 bp) COL4A5 transcripts showing skipping of exon 48. The table displays the relative expression levels of the wild-type and mutated (mut) transcripts determined by densitometry of the agarose gel bands. M, molecular weight marker. (b) Characterization of the mutation in genomic DNA (c.4688G>A).
Transcriptional analysis of patterns of X chromosome inactivation detected a random X inactivation in the hair roots from the proband and in all three tissues analysed from his grandmother (Figure 3).
Determination of the X chromosome inactivation status. Amplification of the AR gene from genomic DNA is shown in panels (a) grandmother and (e) proband, and is performed in order to correct preferential amplification of the smaller allele (allele 1). Expression of both AR alleles is displayed in (b) blood, (c) hair roots and (d) buccal mucosa from the grandmother and in the (f) hair roots from the proband. (g) Quantification of the relative expression levels of both AR alleles showing a random X chromosome inactivation in all analysed samples.
Cytogenetic analysis revealed a normal karyotype in both parents (46,XX and 46,XY) and corroborated the numerical chromosome aberration in the proband: 47,XXY (KS).
Sperm FISH analysis was performed in a total of 181 417 sperm nuclei from the control group (>10 000 sperm/donor) and in 7378 sperm nuclei from the father of the proband, due to his low sperm concentration (2.7 × 106/ml). Disomy rates for chromosomes 6, 21, X and Y and diploidy rates are summarized in Table 1. Only mean values of the control group are displayed because no significant differences among these individuals were found (data not shown). XY disomy was significantly different (P<0.007) in the father of the 47,XXY patient compared to control donors.
Discussion
The first step in the molecular diagnosis of AS is to discern between the X-linked and the autosomal forms of the disease. At first glance, the presence of male-to-male transmission in an AS pedigree discards both X-linked and autosomal recessive forms of the disease. On the other hand, when early ESRD onset is present, autosomal dominant AS is unlikely. The family described here did not fulfil any of these inheritance patterns, thus, we performed linkage analysis for all possible loci. Linkage to COL4A5 detected an extra X chromosome of paternal origin in the proband giving rise to the concurrence of X-linked AS and KS. The 47,XXY karyotype has been identified in males affected by several X-linked diseases such as Fragile X syndrome,11 Rett syndrome,12 Incontinentia pigmenti,13 or Becker muscular dystrophy.14 Nevertheless, the association between AS and KS has not been previously reported. Taking into account that their prevalences are around one in 5000 and one in 1000 male births, respectively, the probability for the simultaneous occurrence of these two disorders is approximately one in 5 000 000 births.
Since KS of paternal origin is the consequence of oocyte fertilization by a disomic sperm, FISH analysis was performed in the father of the proband confirming a significantly increased sperm XY disomy. This increment is the result of a nondisjunction process during meiosis I.6 This is not the first time that a production of aneuploid embryos together with an increment of aneuploid sperm in the father has been detected.15, 16, 17 Moreover, the father of the proband presented with oligozoospermia, which has previously been related to augmented frequencies of sex chromosome disomies and diploidies.18 In addition, an increased rate of disomy 21 has been found in infertile patients.19 We did not observe any significant increase in diploidy, nor in disomy XX, disomy YY or disomy 21. It seems that meiosis I nondisjunction, in this case, is limited to the XY chromosome pair, probably because this bivalent is the most prone to nondisjunction in male gametogenesis.19
No correlation was reported between the severity of the Klinefelter phenotype and origin of the extra X chromosome;20 thus, we cannot predict the KS clinical outcome of this young boy after puberty. However, the early diagnosis of KS in this patient can be considered exceptional since most KS males are not identified until adulthood and it will be extremely helpful in order to anticipate testosterone replacement therapy if necessary. Moreover, cryopreservation of semen samples of this boy in early puberty might preserve his future fertility.
The fact that X-linked AS female carriers have a variable phenotype has led to the hypothesis that part of this clinical heterogeneity could be explained by different patterns of X chromosome inactivation. Guo et al21 reported a severe phenotype in a woman with X-linked AS and demonstrated over 90% inactivation of the wild-type allele in both white blood cells and renal parenchyma. In contrast, no correlation between the severity of the disease and the X-inactivation pattern in white blood cells was found in 43 AS female carriers22 and in 23 others studied by our group (unpublished results). Here we analysed this pattern in three accessible tissues of different embryological origin from the grandmother (blood, hair root and buccal mucosa epithelia) and in the hair root cells of the proband. We detected a random X inactivation in all of them that could allow us to extrapolate a random X inactivation in the AS target tissues. These results were not able to clarify the cause of the severe AS phenotype at 5 years of age in this 47,XXY boy.
The COL4A5 mutation (c.4688G>A) identified in this family is one of the previously described mutations that has been detected in the COL4A5 gene.23, 24 It produces an exon 48 skipping that will yield a truncated polypeptide lacking the noncollagenous domain. Since the c.4688G>A alteration is theoretically a missense substitution (R1563Q), it highlights that mutations should be studied at both the genomic and the RNA level so as to clarify their primary effect on mRNA processing. Interestingly, a preferential expression of the aberrantly spliced transcript was detected in the proband when compared to his grandmother. This finding could explain why the AS manifestations of this boy, with two X chromosomes, resembles more an X-linked AS male than an X-linked AS female carrier. Differences in the RNA splicing machinery between these two patients carrying the same COL4A5 splicing mutation might be responsible for their different levels of exon 48 skipping. Splicing mutations that do not affect the invariant splicing donor GT and acceptor AG intronic dinucleotides, such as the mutation detected in this family, can lead to partial generation of correctly spliced transcripts, which has been associated with phenotypic variability even among siblings. This phenomenon has been previously described in other inherited diseases.25
In conclusion, it is important to stress that when performing molecular diagnosis of AS, the existence of male-to-male transmission must not completely exclude X-linked AS. On the other hand, it has been widely suggested that the variable phenotype in X-linked AS carriers is attributable to different degrees of X chromosome inactivation. However, in the family studied here, this hypothesis has been ruled out and different levels of the aberrantly spliced transcript might explain the differences in disease severity. Further studies should be performed on families with several female carriers bearing a COL4A5 splicing mutation to analyse whether levels of aberrantly spliced transcripts correlate with disease severity.
References
Atkin CL, Gregory MC, Border WA : Alport syndrome; in Schrier RW GC (ed): Diseases of the kidney. Little Brown, Boston, 1988, pp 617–641.
Barker DF, Hostikka SL, Zhou J et al: Identification of mutations in the COL4A5 collagen gene in Alport syndrome. Science 1990; 248: 1224–1227.
Mochizuki T, Lemmink HH, Mariyama M et al: Identification of mutations in the alpha 3(IV) and alpha 4(IV) collagen genes in autosomal recessive Alport syndrome. Nat Genet 1994; 8: 77–81.
Jefferson JA, Lemmink HH, Hughes AE et al: Autosomal dominant Alport syndrome linked to the type IV collagen alpha 3 and alpha 4 genes (COL4A3 and COL4A4). Nephrol Dial Transplant 1997; 12: 1595–1599.
Lanfranco F, Kamischke A, Zitzmann M, Nieschlag E : Klinefelter's syndrome. Lancet 2004; 364: 273–283.
Thomas NS, Hassold TJ : Aberrant recombination and the origin of Klinefelter syndrome. Hum Reprod Update 2003; 9: 309–317.
Renieri A, Bruttini M, Galli L et al: X-linked Alport syndrome: an SSCP-based mutation survey over all 51 exons of the COL4A5 gene. Am J Hum Genet 1996; 58: 1192–1204.
Dunning AM, McBride S, Gregory J et al: No association between androgen or vitamin D receptor gene polymorphisms and risk of breast cancer. Carcinogenesis 1999; 20: 2131–2135.
Lau AW, Brown CJ, Penaherrera M, Langlois S, Kalousek DK, Robinson WP : Skewed X-chromosome inactivation is common in fetuses or newborns associated with confined placental mosaicism. Am J Hum Genet 1997; 61: 1353–1361.
Arnedo N, Nogues C, Bosch M, Templado C : Mitotic and meiotic behaviour of a naturally transmitted ring Y chromosome: reproductive risk evaluation. Hum Reprod 2005; 20: 462–468.
Santos CB, Hjalgrim H, Carneiro FR, Ribeiro M, Boy RT, Pimentel MM : Concurrence of fragile X and Klinefelter syndromes: report of a new case of paternal nondisjunction. Ann Genet 2003; 46: 53–55.
Schwartzman JS, Bernardino A, Nishimura A, Gomes RR, Zatz M : Rett syndrome in a boy with a 47,XXY karyotype confirmed by a rare mutation in the MECP2 gene. Neuropediatrics 2001; 32: 162–164.
Kenwrick S, Woffendin H, Jakins T et al: Survival of male patients with incontinentia pigmenti carrying a lethal mutation can be explained by somatic mosaicism or Klinefelter syndrome. Am J Hum Genet 2001; 69: 1210–1217.
Suthers GK, Manson JI, Stern LM, Haan EA, Mulley JC : Becker muscular dystrophy (BMD) and Klinefelter's syndrome: a possible cause of variable expression of BMD within a pedigree. J Med Genet 1989; 26: 251–254.
Blanco J, Gabau E, Gomez D et al: Chromosome 21 disomy in the spermatozoa of the fathers of children with trisomy 21, in a population with a high prevalence of Down syndrome: increased incidence in cases of paternal origin. Am J Hum Genet 1998; 63: 1067–1072.
Martinez-Pasarell O, Nogues C, Bosch M, Egozcue J, Templado C : Analysis of sex chromosome aneuploidy in sperm from fathers of Turner syndrome patients. Hum Genet 1999; 104: 345–349.
Eskenazi B, Wyrobek AJ, Kidd SA et al: Sperm aneuploidy in fathers of children with paternally and maternally inherited Klinefelter syndrome. Hum Reprod 2002; 17: 576–583.
Martin RH, Rademaker AW, Greene C et al: A comparison of the frequency of sperm chromosome abnormalities in men with mild, moderate, and severe oligozoospermia. Biol Reprod 2003; 69: 535–539.
Tempest HG, Griffin DK : The relationship between male infertility and increased levels of sperm disomy. Cytogenet Genome Res 2004; 107: 83–94.
Iitsuka Y, Bock A, Nguyen DD, Samango-Sprouse CA, Simpson JL, Bischoff FZ : Evidence of skewed X-chromosome inactivation in 47,XXY and 48,XXYY Klinefelter patients. Am J Med Genet 2001; 98: 25–31.
Guo C, Van Damme B, Vanrenterghem Y, Devriendt K, Cassiman JJ, Marynen P : Severe alport phenotype in a woman with two missense mutations in the same COL4A5 gene and preponderant inactivation of the X chromosome carrying the normal allele. J Clin Invest 1995; 95: 1832–1837.
Vetrie D, Flinter F, Bobrow M, Harris A : X inactivation patterns in females with Alport's syndrome: a means of selecting against a deleterious gene? J Med Genet 1992; 29: 663–666.
Zhou J, Gregory MC, Hertz JM et al: Mutations in the codon for a conserved arginine-1563 in the COL4A5 collagen gene in Alport syndrome. Kidney Int 1993; 43: 722–729.
Lemmink HH, Kluijtmans LA, Brunner HG et al: Aberrant splicing of the COL4A5 gene in patients with Alport syndrome. Hum Mol Genet 1994; 3: 317–322.
Nissim-Rafinia M, Kerem B : Splicing regulation as a potential genetic modifier. Trends Genet 2002; 18: 123–127.
Acknowledgements
We thank the family for taking part in this study. We also thank Dr Moisès Burset and Helena Kruyer for correction of the manuscript. This work was supported by grants of the Ministerio de Sanidad (02/3672, Infraestructura 2002), the Red de Enfermedades Metabólicas Hereditarias (03/054), the Ministerio de Ciencia y Tecnología (BFI2002-01193) and the Generalitat de Catalunya (2001SGR-00202), Spain.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ars, E., Tazón-Vega, B., Ruiz, P. et al. Male-to-male transmission of X-linked Alport syndrome in a boy with a 47,XXY karyotype. Eur J Hum Genet 13, 1040–1046 (2005). https://doi.org/10.1038/sj.ejhg.5201452
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5201452
Keywords
This article is cited by
-
Targeted next-generation sequencing in steroid-resistant nephrotic syndrome: mutations in multiple glomerular genes may influence disease severity
European Journal of Human Genetics (2015)