Abstract
Osteoporosis is a common condition characterized by reduced skeletal strength and increased susceptibility to fracture. The single major risk factor for osteoporosis is low bone mineral density (BMD) and strong evidence exists that genetic factors are in part responsible for an individual's BMD. A cohort of 40 multiplex Caucasian families selected through a proband with osteoporosis was genotyped for microsatellite markers spaced at an average of 10 cM, and linkage to femoral neck (FN), lumbar spine (LS) and trochanter (TR) BMD was analyzed using univariate and bivariate variance component linkage analysis. Maximum univariate multipoint lod-scores were 2.87 on chromosome 1p36 for FN BMD, 1.89 on 6q27 for TR BMD, and 2.15 on 7p15 for LS BMD. Results of bivariate linkage analysis were highly correlated with those of the univariate analysis, although generally less significant, suggesting the possibility that some of these susceptibility loci may exert pleiotropic effects on multiple skeletal sites.
Similar content being viewed by others
Introduction
Osteoporosis is a systemic skeletal disease characterized by low bone mass and deterioration of bone microarchitecture that result in bone fragility and susceptibility to fracture.1 Positive family history is one of the major risk factors for osteoporosis, and epidemiological studies indicate that genetic factors contribute substantially to susceptibility to osteoporosis.2
In 1998 we first reported a whole-genome scan of osteoporosis-related traits, based on the analysis of a small sample of extended Caucasian pedigrees of Canadian origin that had been selected through a proband with low bone mineral density (BMD).3 Our study focused on quantitative trait linkage analysis of lumbar spine (LS) and femoral neck (FN) BMD and identified three regions with lod-scores ⩾2.2 on chromosomes 1p, 2p, and 4q. Linkage of FN BMD to chromosome 1p36 was confirmed by high-density mapping in an extended sample that included 36 additional families selected from the same population.4
Since our initial study, several other groups have reported genome scans for bone-related traits (reviewed in Liu et al;5 also more recently Kammerer et al,6 Styrkarsdottir et al,7 Wilson et al,8 Huang et al,9 Karasik et al,10 and Shen et al11). In spite of heterogeneity in sampling strategies, statistical approaches, traits, and populations, these studies have identified several common chromosomal regions that are candidates for loci responsible for susceptibility to osteoporosis.12 Among these regions, chromosome 1p36, initially identified in our pedigrees3, 4 has been confirmed to contain a quantitative trait locus (QTL) for bone-related traits in several other studies.8, 13, 14 As in the analysis of all complex traits, replication in independent studies is a key parameter in defining the significance of a linkage finding.
We have now extended the analysis of the whole autosomal genome to our entire collection of osteoporosis families and added the evaluation of trochanter (TR) BMD in addition to LS and FN BMD. Here, we report the results of univariate and bivariate variance component (VC) linkage analysis to identify QTLs for bone density that may affect risk of osteoporosis in this and possibly other groups of families.
Materials and methods
Families and phenotypes
The 40 families used in this analysis were part of a larger group described previously.3, 4 They included six multigenerational families, 28 nuclear families with two to four children and six three-generation families with two sibships of two to four offspring. Two families of two sisters each without parents that were included in a previous study4 have been excluded from the present study because X-chromosome microsatellite marker genotypes were inconsistent with a sibling relationship.
All families were identified and recruited at Montreal General Hospital through the presence of a proband who had a LS BMD Z-score of ⩽−2.00 as measured by dual energy X-ray absorptiometry (DEXA) using the Lunar DPX densitometer (Lunar, Madison WI, USA), and who reported having relatives who might also be affected with osteoporosis. All available relatives 18 years old or older were examined at Montreal General Hospital, and bone density determinations at the LS (L2–L4), the FN, and the TR were made on the same instrument. All those with known causes (other than estrogen depletion due to menopause) of metabolic bone disease were eliminated from further study. A 5–15 ml blood sample was collected from each participating individual for either genomic DNA extraction or lymphocyte culture. All subjects gave written informed consent, and the study was approved by the Institutional Review Board of the participating institutions.
Genotyping
DNA was extracted from blood directly or from lymphoblast cultures established from patients' lymphocytes by EBV transformation. Microsatellite marker genotyping was begun in the laboratory of Dr Darwin Prockop (chromosomes 1–6, 11, 19–22) using ABI Prism Linkage Mapping Set Version 2.0 and 2.5 and completed in the laboratory of Dr Katia Sol-Church (chromosomes 7–10, 12–18) using ABI Prism Linkage Mapping Set Version 2.5. Using multicolor fluorescent dye technology, chromosome-specific microsatellite markers were amplified under single or multiplex PCR conditions. The reactions were pooled either manually or using a Biomek 2000, combined with a size standard and loaded on a ABI Prism 377 and separated by gel electrophoresis. GeneScan 3.1 software was used to size the specific PCR products. Genotyper 2.1 was used for allele calling prior to statistical analysis.
Statistical genetic analysis
All statistical genetic analyses except the multipoint identity by descent (IBD) calculation were performed using the computer software SOLAR 2.1.2 (http://www.sfbr.org/solar/index.html).15
We first estimated heritability of LS, FN, and TR BMD in our sample of families as the proportion of the trait variance attributable to additive genetic factors, while simultaneously estimating the significance of the effect of the covariates sex, age, age squared, sex-specific age (or sex × age interaction), body mass index (BMI), and ethnicity. All covariates with associated P-values of 0.1 or less from the quantitative analysis were retained in the subsequent analyses. Genetic and environmental correlations between pairs of traits and their significance were also estimated using a maximum likelihood procedure.
Univariate multipoint linkage analysis of BMD to microsatellite markers was carried out using the VC approach separately for all three traits. In addition, to evaluate the hypothesis that common genes may determine bone density variation at the three sites, we performed bivariate VC linkage analysis on microsatellite markers for all chromosomes. Bivariate linkage analysis has been shown to increase power to detect linkage of related traits to a common QTL as it exploits the additional information contained in the correlation pattern between the two quantitative traits.16 However, the lod-scores thus obtained cannot be directly compared to the univariate lod-scores as bivariate lod-scores involve two degrees of freedom. Instead, asymptotic P-values for the bivariate lod-scores can be calculated based on a 1/4X22:1/2X21:1/4X20 distribution for the 2 ln(10) lod transformation.16
In the VC approach, the total trait phenotypic variance is partitioned into components attributable to the effects of the covariates, the effect of the specific locus under scrutiny (QTL), and residual additive genetic effects. Prior to the linkage analysis, the proportion of alleles shared IBD by pairs of relatives was estimated using the Markov chain MonteCarlo algorithm implemented in the computer program Simwalk2 version 2.86 (http://watson.hgen.pitt.edu/docs/simwalk2.html)17 at each marker location and every 1 cM between adjacent markers along any given chromosome. In contrast to the multipoint procedure implemented in SOLAR, which is based on interpolation between single point values, this is a true multipoint IBD calculation, and thus more fully extracts all genetic information contained in the microsatellite marker data. These IBD probabilities were then imported into SOLAR and used to estimate the genetic variance attributable to a hypothetical QTL linked to any given location. A test for linkage was carried out by testing whether the QTL variance was significantly different from 0 by comparing the likelihood of this model with that of a restricted model in which the genetic variance at the same location was fixed at 0.
To account for possible deviations of the trait distributions from the assumption of multivariate normality, which may inflate the probability of type I error, we estimated the significance of the maximum lod-scores that we observed by means of an empirical null distribution obtained by MonteCarlo simulation of 10 000 replicates of an unlinked marker.
As families were ascertained through the presence of an individual with low LS BMD, all LS univariate linkage analyses were repeated including an ascertainment correction by conditioning on the likelihood of observing the BMD value of the proband. Differences observed between the results of the two kinds of analyses were trivial, with lod-scores differing by at most 0.1, and only results without the ascertainment correction are reported here for consistency with the other traits.
Marker allele frequencies used in all linkage analyses were estimated in our families by maximum likelihood. Marker order and genetic distances used in the multipoint linkage analysis were as reported in the sex-average genetic maps of the Marshfield Center for Medical Genetics (http://research.marshfieldclinic.org/genetics/). Physical locations for markers and genes are from Ensembl v.26 (http://www.ensembl.org/) and are based on Human NCBI build 35.
Results
Heritabilities were estimated at 0.84 for LS BMD (standard error (SE): 0.09, P<0.0001), 0.52 for FN BMD (SE: 0.13, P<0.0001), and 0.55 for TR BMD (SE: 0.14, P<0.0001). Sex (P-values <0.000001 for FN and TR, and 0.05 for LS) and BMI (all P-values <0.001) significantly contributed to variation in all three traits; a first-order age effect was detected for LS and FN (P-values <0.01) and a significant age × sex interaction was observed for LS and TR BMD (P-values <0.01).
Bivariate genetic analysis indicated that significant genetic and environmental correlation exists between all pairs of the three traits (Table 1), supporting the hypothesis that shared genetic as well as environmental factors account for a significant portion of the total trait variance. However, tests for pleiotropy rejected the hypothesis that the genetic correlations were equal to 1, thus indicating that unique nonshared genes also significantly contribute to each trait variation.
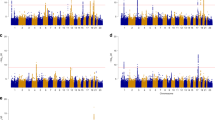
A summary of the maximum lod-scores observed for the three traits on each chromosome in the univariate VC linkage analysis is reported in Table 2, and Figure 1 shows the univariate multipoint graphs for all chromosomes where we observed lod-scores greater than 1. The highest multipoint lod-score was 2.87 for FN BMD at 16 cM, close to marker D1S2694 which is located at 7.2 Mb on 1p36.3. The empirical P-value associated with this lod-score was estimated at 0.0003 based on 10 000 replicates of an unlinked marker. For the other two traits, the maximum lod-scores observed in 1p36 were 0.68 (LS BMD), and 1.11 (TR BMD) both at 15 cM.
The genome-wide maximum multipoint lod-score for LS BMD was 2.15 on chromosome 7p15 at 34 cM, close to D7S493 (empirical P-value=0.003). For TR BMD, the genome-wide maximum multipoint lod-score was 1.89 on chromosome 6q27 at 189 cM, close to D6S446 (empirical P-value=0.002).
Additional lod-scores >1 were observed for FN BMD at 2p23, 5q33, 12q24, and 22q12; for LS BMD at 1q21, 4q35, 5q33, and 22q12; and for TR BMD at 1q32, 7p22, and 12q24 (Table 2).
Results of bivariate VC linkage analysis are reported in Figure 2 for those chromosomes where we observed lod-scores >1.4 (roughly equivalent to univariate lod-scores of >1) and summarized in Table 3 for all the chromosomes. The maximum lod-score we observed was 2.55 (asymptotic P-value=0.001) for FN and LS BMD near the location of the maximum univariate lod-score in 1p36. Bivariate analysis of FN and TR BMD also produced a maximum lod-score greater than 2 at the same location (Zmax=2.01, asymptotic P-value=0.004).
A maximum lod-score of 2.00 (asymptotic P-value=0.004) was also obtained for bivariate analysis of FN and LS BMD near marker D4S1535 on 4q35. All other lod-scores from the bivariate analysis were less than 2.
Discussion
Chromosome 1p36 continues to have the strongest support for a locus that contributes to bone density in this whole-genome scan of the entire cohort of 40 families collected through a proband with low spinal BMD. The support is from both univariate and bivariate VC linkage analysis, with the maximum lod-score of 2.87 near marker D1S2694 for the FN BMD trait. In addition, the lod-scores from bivariate analysis are 2.55 and 2.01 for FN/LS BMD and FN/TR BMD, respectively. We previously reported a maximum lod-score of 3.5 for FN BMD at marker D1S214, approximately 1 cM away from D1S2694, in our fine mapping of the chromosome 1p36 region.4 The difference between the two univariate linkage analysis results can be explained by the lower marker density used in the present study (only markers included in the ABI Prism Linkage Mapping Set Version 2.5 were used for consistency with the other chromosomes), the different multipoint procedure employed, and the fact that two small families were dropped in the present study. The finding of bivariate lod-scores >2 for the same region for the FN/LS BMD and FN/TR BMD trait pairs suggests that the same QTL may exert an effect on bone density at all three skeletal sites tested in our families. Although the bivariate lod-scores were less significant than the maximum lod-score we observed for FN BMD alone, they were higher than those obtained for LS BMD and TR BMD separately. This suggests that identification of linkage of these traits to this region benefits from the increase in power resulting from the use of bivariate linkage analysis.
A number of independent studies have reported linkage or association of bone-related traits to markers of chromosome 1p36.8, 13, 14 In addition, different groups have reported linkage of mouse chromosome 4q, the homologous region to human 1p, to bone density.18, 19, 20 Taken together, all these findings provide convincing evidence for the existence of a bone-related QTL in the distal part of chromosome 1p. The observation of linkage to 1p36 from our univariate analysis of FN BMD, and bivariate analysis of FN/LS BMD and FN/TR BMD, and from other studies of different measurements of bone density (such as total body BMD8 and quantitative ultrasound of the calcaneus bone13) suggests that this QTL may influence variation of BMD in general rather than at a specific skeletal site.
The highest lod-score we observed for LS BMD was 2.15 and occurred on chromosome 7p15 at marker D7S493 (located at 21.5 Mb) close to IL-6 (located at 22.5 Mb). Maximum bivariate lod-scores in the same region were 1.59 for FN/LS BMD and 1.96 for LS/TR BMD. Several studies have reported association of polymorphisms of IL-6, and in particular of the promoter G-174C polymorphism, with BMD or other bone-related traits, mostly in postmenopausal women.12 Another interesting gene in this region is the human osteoactivin gene located at 23 Mb. Osteoactivin (OA) is highly expressed in the bones of osteopetrotic mutant rats.21 Human and mouse OA are expressed in primary osteoblast cultures at all stages of differentiation.22
The highest univariate lod-score for TR BMD was 1.89 on chromosome 6q27 near marker D6S446 at 170 Mb. In this case, the bivariate analysis of trait pairs LS/TR BMD gave a maximum lod-score of 1.75 in the same area. To the best of our knowledge, there are no previously reported BMD candidate genes in this region of the genome. Inspection of the human genome sequence around D6S446 did not reveal any obvious candidate gene. The estrogen receptor 1 (ESR1) that has been extensively investigated in association to BMD and other osteoporosis related traits12 is located on chromosome 6q25 at 152.2 Mb and as such is unlikely to be a candidate for this putative QTL.
The two other regions where we previously reported lod-scores greater than 2 were 2p23–p24 and 4q32–q34.3 The lod-scores in both these regions are now lower than those we previously reported in a subset of these families.3 In the present study, chromosome 4q35 yielded a lod-score of 1.3 for LS BMD near D4S1535, and bivariate lod-scores of 2.00 and 1.16 for the FN/LS BMD and LS/TR BMD trait pairs.
In our first genome scan, chromosome 2p23 gave a multipoint lod score of 2.29 for LS BMD, but in this study gave a lod-score of 1.53 (near marker D2S165) for FN BMD and bivariate lod-scores >1 for FN/LS BMD and FN/TR BMD. Marker D2S405 in the same region on 2p23 was reported to be linked to forearm BMD by Niu et al.23
Other regions where we observed interesting lod-scores coincide with regions reported in other studies and/or known to contain candidate genes for bone-related traits.5, 12 A formal meta-analysis will help elucidate whether these findings may in fact indicate the existence of genomic regions containing susceptibility genes for osteoporosis shared across different populations. In particular, chromosome 5q33 yielded lod-scores of more than 1 in both univariate analysis for FN BMD and LS BMD and bivariate analysis for FN/LS BMD and FN/TR BMD in our pedigrees, and coincides with the region reported by Koller et al24 for FN BMD. This region contains the osteonectin (ON) and the platelet-derived growth factor receptor beta (PDGFRB) genes, both of which have been previously suggested as candidate gene for BMD.24
Kammerer et al6 reported linkage of forearm BMD at marker D12S2070, which is located at 114.5 Mb on chromosome 12q24 approximately 22 kb away from marker D12S79, where we found lod-scores of >1 for both FN and TR BMD. Karasik et al25 also found linkage of LS BMD near marker D12S395, located at 118.6 Mb on chromosome 12q24, whereas Deng et al26 found linkage of LS near D12S1723, located at 130 Mb. All together, these results suggest the presence of a locus with pleiotropic effects on BMD at different skeletal sites in 12q24. However, our own bivariate linkage analysis failed to detect a strong signal to this region.
In addition to the 1p36 peak for FN BMD, two smaller peaks were observed on chromosome 1q for LS BMD at 153 cM (Zmax=1.43), and for TR BMD at 219 cM (Zmax=1.29). The bivariate Zmax in 1q for the same two traits was 1.69 at 150 cM. Econs et al27 have recently confirmed linkage of LS BMD to 1q in a large sister-pair study, with a peak at 181 cM in the Marshfield genetic map. Given the difficulty in precisely locating genes responsible for complex traits by linkage methods, we cannot exclude the possibility that these different peaks represent the same QTL.
In this study, we used bivariate in addition to univariate linkage analysis to test whether the BMD traits we analyzed may share QTLs at any of the tested positions in the genome. Bivariate linkage analysis has been shown to have increased power over univariate analysis in detecting genes of pleiotropic effect.16 A significant pleiotropic effect on LS and FN BMD was estimated in a large sample from an intercontinental study,28 and our own quantitative analysis showed that the genetic correlations at all three pairs of skeletal sites are significantly different from 0 (Table 1). Others10, 29 have used principal component analysis to test for common QTLs underlying bone mass at different skeletal sites. Results of our bivariate linkage analysis are generally correlated with those of the univariate analysis, with the same genomic regions producing lod-scores >1 in both univariate and bivariate analyses, suggesting that if indeed there are BMD QTLs located in these regions they may exert an effect on BMD at multiple sites in the skeleton (Tables 2 and 3). A notable exception is chromosome 22q12 for FN and LS which gave lod-scores >1 in the univariate analysis but not in the bivariate analysis. However, bivariate lod-scores were usually less significant than the maximum univariate lod-score at the same location. For example, the maximum bivariate lod-scores at 1p36 for FN/LS BMD (2.55) and FN/TR BMD (2.01) are lower than the maximum lod-score for FN BMD (2.87), but higher than those for LS BMD (0.68) and TR BMD (1.11) in the same region. This possibly reflects a difference in effect exerted by the putative QTL at the different skeletal sites. While the effect on FN BMD was strong enough to be detected by the univariate analysis, the bivariate analysis with FN BMD allowed detection of a weaker effect for LS BMD and TR BMD that was not evident from the univariate analysis. In addition, it has been shown that in order to gain from the increase in power of multivariate linkage analysis, the genetic correlation at the QTL and the residual correlation must be of different sign.30, 31 In particular, a larger increase is observed when the genetic correlation due to the QTL is negative and the residual correlation is positive.31 However, all the correlations estimated at the 1p36 QTL among our traits were in fact positive.
In conclusion, a whole autosomal genome scan supports 1p36 as the location of the major QTL for BMD in our families. Given the results of our univariate and bivariate linkage analyses, and those reported from other independent studies,8, 13, 14 it is likely that this represents a gene responsible for controlling BMD at several skeletal sites. However, as one can expect for a complex phenotype such as BMD, several other loci of a less significant effect may also be present in other regions of the genome. Importantly, these other loci may be located in regions of the genome that coincide with those reported in other linkage studies or known to contain promising candidate genes for controlling BMD. These findings support further analysis of these candidate genomic regions and genes in these and other families.
References
Consensus development conference: diagnosis, prophylaxis, and treatment of osteoporosis. Am J Med 1993; 94: 646–650.
Stewart TL, Ralston SH : Role of genetic factors in the pathogenesis of osteoporosis. J Endocrinol 2000; 166: 235–245.
Devoto M, Shimoya K, Caminis J et al: A first-stage autosomal genome screen in extended pedigrees suggests genes predisposing to low bone mineral density are on chromosome 1p, 2p, and 4q. Eur J Hum Genet 1998; 6: 151–157.
Devoto M, Specchia C, Li HH et al: Variance component linkage analysis indicates a QTL for femoral neck bone mineral density on chromosome 1p36. Hum Mol Genet 2001; 10: 2447–2452.
Liu YZ, Liu YJ, Recker RR, Deng HW : Molecular studies of identification of genes for osteoporosis: the 2002 update. J Endocrinol 2003; 177: 147–196.
Kammerer CM, Schneider JL, Cole SA et al: Quantitative trait loci on chromosomes 2p, 4p, and 13q influence bone mineral density of the forearm and hip in Mexican Americans. J Bone Miner Res 2003; 18: 2245–2252.
Styrkarsdottir U, Cazier JB, Kong A et al: Linkage of osteoporosis to chromosome 20p12 and association to BMP2. PLoS Biol 2003; 1: E69.
Wilson SG, Reed PW, Bansal A et al: Comparison of genome screens for two independent cohorts provides replication of suggestive linkage of bone mineral density to 3p21 and 1p36. Am J Hum Genet 2003; 72: 144–155.
Huang QY, Xu FH, Shen H et al: A second-stage genome scan for QTLs influencing BMD variation. Calcif Tissue Int 2004; 75: 138–143.
Karasik D, Cupples LA, Hannan MT, Kiel DP : Genome screen for a combined bone phenotype using principal component analysis: the Framingham study. Bone 2004; 34: 547–556.
Shen H, Zhang YY, Long JR et al: A genome-wide linkage scan for bone mineral density in an extended sample: evidence for linkage on 11q23 and Xq27. J Med Genet 2004; 41: 743–751.
Huang QY, Recker RR, Deng HW : Searching for osteoporosis genes in the post-genome era: progress and challenges. Osteoporos Int 2003; 14: 701–715.
Karasik D, Myers RH, Hannan MT et al: Mapping of quantitative ultrasound of the calcaneus bone to chromosome 1 by genome-wide linkage analysis. Osteoporos Int 2002; 13: 796–802.
Wynne F, Drummond FJ, Daly M et al: Suggestive linkage of 2p22–25 and 11q12–13 with low bone mineral density at the lumbar spine in the Irish population. Calcif Tissue Int 2003; 72: 651–658.
Almasy L, Blangero J : Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet 1998; 62: 1198–1211.
Almasy L, Dyer TD, Blangero J : Bivariate quantitative trait linkage analysis: pleiotropy versus co-incident linkages. Genet Epidemiol 1997; 14: 953–958.
Sobel E, Lange K : Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker sharing statistics. Am J Hum Genet 1996; 58: 1323–1337.
Beamer WG, Shultz KL, Donahue LR et al: Quantitative trait loci for femoral and lumbar vertebral bone mineral density in C57BL/6J and C3H/HeJ inbred strains of mice. J Bone Miner Res 2001; 16: 1195–1206.
Klein OF, Carlos AS, Vartanian KA et al: Confirmation and fine mapping of chromosomal regions influencing peak bone mass in mice. J Bone Miner Res 2001; 16: 1953–1961.
Masinde GL, Li X, Gu W, Wergedal J, Mohan S, Baylink DJ : Quantitative trait loci for bone density in mice: the genes determining total skeletal density and femur density show little overlap in F2 mice. Calcif Tissue Int 2003; 71: 421–428.
Safadi FF, Xu J, Smock SL, Rico MC, Owen TA, Popoff SN : Cloning and characterization of osteoactivin, a novel cDNA expressed in osteoblasts. J Cell Biochem 2001; 84: 12–26.
Owen TA, Smock SL, Prakash S et al: Identification and characterization of the genes encoding human and mouse osteoactivin. Crit Rev Eukaryot Gene Expr 2003; 13: 205–220.
Niu T, Chen C, Cordell H et al: A genome-wide scan for loci linked to forearm bone mineral density. Hum Genet 1999; 104: 226–233.
Koller DL, Econs MJ, Morin PA et al: Genome screen for QTLs contributing to normal variation in bone mineral density and osteoporosis. J Clin Endocrinol Metab 2000; 85: 3116–3120.
Karasik D, Myers RH, Cupples LA et al: Genome screen for quantitative trait loci contributing to normal variation in bone mineral density: the Framingham study. J Bone Miner Res 2002; 17: 1718–1727.
Deng HW, Xu FH, Huang QY et al: A whole genome scan suggests several genomic regions potentially containing quantitative trait loci for osteoporosis. J Clin Endocrinol Metab 2002; 87: 5151–5159.
Econs MJ, Koller DL, Hui SL et al: Confirmation of linkage to chromosome 1q for peak vertebral bone mineral density in premenopausal white women. Am J Hum Genet 2004; 74: 223–228.
Livshits G, Deng HW, Nguyen TV, Yakovenko K, Recker RR, Eisman JA : Genetics of bone mineral density: evidence for a major pleiotropic effect from an intercontinental study. J Bone Min Res 2004; 19: 914–923.
Peacock M, Koller DL, Hui S, Johnston CC, Foroud T, Econs MJ : Peak bone mineral density at the hip is linked to chromosomes 14q and 15q. Osteoporos Int 2004; 15: 489–496.
Allison DB, Thiel B, St Jean P, Elston RC, Infante MC, Schork NJ : Multiple phenotype modeling in gene-mapping studies of quantitative traits: power advantages. Am J Hum Genet 1998; 63: 1190–1201.
Amos C, de Andrade M, Zhu D : Comparison of multivariate tests for genetic linkage. Hum Hered 2001; 51: 133–144.
Acknowledgements
This work was supported by grants HD36606 (LDS) and 38188 (DP) from NIH, and by grant 2001-097 from Nemours Biomedical Research (MD).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Devoto, M., Spotila, L., Stabley, D. et al. Univariate and bivariate variance component linkage analysis of a whole-genome scan for loci contributing to bone mineral density. Eur J Hum Genet 13, 781–788 (2005). https://doi.org/10.1038/sj.ejhg.5201411
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5201411
Keywords
This article is cited by
-
Evaluation of common variants in CNR2 gene for bone mineral density and osteoporosis susceptibility in postmenopausal women of Han Chinese
Osteoporosis International (2015)
-
Bone mineral density is linked to 1p36 and 7p15-13 in a southern Chinese population
Journal of Bone and Mineral Metabolism (2011)
-
High resolution linkage and linkage disequilibrium analyses of chromosome 1p36 SNPs identify new positional candidate genes for low bone mineral density
Osteoporosis International (2009)
-
Cannabinoid receptors and the regulation of bone mass
British Journal of Pharmacology (2008)
-
Bone structural effects of variation in the TNFRSF1B gene encoding the tumor necrosis factor receptor 2
Osteoporosis International (2008)