Abstract
We report a family heterozygous for a newly identified mutation in the tyrosine kinase I domain of the FGFR2 gene (1576A>G, encoding the missense substitution Lys526Glu), associated with variable expressivity of Crouzon syndrome, including clinical nonpenetrance. Our observations expand both the clinical and molecular spectrum of this unusual subset of FGFR2 mutations.
Similar content being viewed by others
Introduction
Crouzon craniofacial dysostosis (MIM 123500) is a well-recognised autosomal dominant craniosynostosis syndrome with a birth prevalence of approximately 1 in 60 000.1 The characteristic facial features are acrocephaly, exorbitism, maxillary hypoplasia, beaked nose and prominent mandible. In contrast to Pfeiffer syndrome (MIM 101600), in which the first digits are characteristically broad and angulated, limb anomalies in Crouzon syndrome are milder or only apparent on radiological examination.2, 3 Heterozygous mutations of the gene encoding fibroblast growth factor receptor type 2 (FGFR2), highly localised to two exons (termed IIIa and IIIc, encoding the extracellular IgIIIa/c domain of the protein), account for the majority of classical cases.4, 5, 6 Recently, we identified a new cluster of relatively rare FGFR2 mutations within the intracellular tyrosine kinase (TK) domain in patients with Crouzon and Pfeiffer syndromes.7 Here we describe a novel TK domain mutation (1576A>G, encoding the missense change Lys526Glu), segregating with a particularly mild Crouzon syndrome phenotype, including nonpenetrance in one individual.
Patients, materials and methods
Patients
The family was initially referred to the Genetic Counselling Clinic in Leuven. Informed consent was given to obtain blood samples for genetic analysis.
Mutation screening
The entire coding region of FGFR2 (GenBank NM_000141; 592 has been subtracted from the reference numbering so that the first nucleotide of the start codon=1) was screened for mutations using denaturing high-performance liquid chromatography on the WAVE Nucleic Acid Fragment Analyser System (Transgenomic Ltd, Crewe, Cheshire, UK) using the primers and methods described previously.7 An altered migration pattern was observed for exon 14 in the proband's DNA, which was sequenced by means of the BigDye terminator kit (Applied Biosystems, Foster City, California, USA) and analysed on the ABI PRISM 3100 sequencer (Applied Biosystems). The sequence change was confirmed by digestion with the restriction enzyme BbsI (New England Biolabs (UK) Ltd, Hitchin, Hertfordshire, UK). The 1576A>G substitution results in cleavage of the 250 bp exon 14 product into fragments of 82 and 168 bp, which was visualised on a 4% Metaphor agarose gel (FMC Bioproducts). Screening of the entire remainder of the FGFR2 gene, including exons IIIa and IIIc, was normal.
Results and discussion
Case report
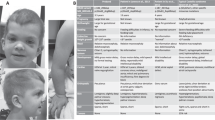
The female proband (Figure 1a) initially presented at the age of 15 years to the Genetic Counselling Clinic for discussion of a clinical diagnosis of Crouzon syndrome. She had exorbitism, marked zygomatic hypoplasia, a beaked nose, a thin upper lip with a short frenulum and relative prognathism. A Le Fort I osteotomy of the maxilla was performed, aged 17 years. At presentation as an adult, her height was 164 cm and head circumference 56.4 cm (within normal limits). She had mild infra-orbital hypoplasia, a short philtrum, a high-arched palate and mild broadening of the thumbs. Psychomotor function was within the normal range.
Her father (Figure 1b), the eighth in a sibship of 9, was born to a 37-year-old father and 32-year-old mother. He commented that several of his siblings had a similar facial appearance, but none was available for clinical assessment. His height was 180 cm and head circumference 60 cm (+3 SD). He had infra-orbital hypoplasia and a high arched palate. He had very mild broadening of the thumbs and great toes, and mild cutaneous syndactyly of the fingers. The clinical diagnosis of Crouzon syndrome was confirmed in the proband and her father, taking into account the craniofacial appearance, only mild broadening of first digits and lack of requirement for calvarial remodelling.
The proband's sister (Figure 1c) had neither facial dysmorphism nor hand or foot anomalies.
Genetic analysis
A heterozygous nucleotide transition, 1576A>G, was initially identified in the proband (Figure 2a) and its presence confirmed in her father and sister by diagnostic restriction enzyme digest with BbsI (Figure 2b). This mutation is predicted to encode an amino-acid substitution from lysine to glutamate at position 526 of the N-terminal lobe of the tyrosine kinase domain (TK1). This is very likely to be the cause of the phenotype in this family because (1) the lysine is highly conserved in vertebrate fibroblast growth factor receptors;8 (2) the substitution is chemically nonconservative (basic to acidic) and is predicted to disrupt a salt bridge with the aspartate side chain at position 530;9 (3) it was not observed in 182 normal control chromosomes. Nevertheless, the associated phenotype is particularly mild, including nonpenetrance in one individual.
Identification of FGFR2 mutation in the family and sequence context in TK1 domain. (a) DNA sequence chromatograms showing the heterozygous 1576A>G transition present in the proband (upper trace) but not in a normal sample (lower trace). (b) Digestion of exon 14 PCR product (U, uncut product) with BbsI reveals a new restriction site generated by the mutation in all three family members, not present in a normal control sample (N). The position of migration (in base pairs) of φX174-HaeIII size standards is indicated to the right. (c) The site of the Lys526Glu substitution (red E) and extent of αC helix (rectangle) are shown above the normal human FGFR2 sequence. Below is an alignment with TK1 domain sequences from human FGFR1, FGFR3, FGFR4; Drosophila melanogaster (HTL, BTL) and Caenorhabditis elegans (EGL-15) FGFR homologues; and the receptor tyrosine kinase proteins KIT, RET and MET. Residues in which missense mutations have been recorded are shaded according to their functional effects based on combined genetic and biochemical evidence: blue, loss of function; green, gain of function (adapted from Kan11).
Comparison with the previously described FGFR2 mutations associated with craniosynostosis7, 10 shows that the Lys526Glu mutation is the most N-terminal mutation in the TK1 domain recorded to date. According to the structure of the paralogous protein FGFR1, the mutation resides in the α-helical segment (termed αC) of TK1.9 Although no mutation of any human FGFR has previously been described in this helix, its position corresponds to a cluster of mutations described in other receptor tyrosine kinase proteins, including KIT, RET and the Caenorhabditis elegans FGFR orthologue EGL-15 (Figure 2c). These mutations may be associated with either loss- or gain-of-function (see legend to Figure 2c), making it difficult to predict the pathological effect of the Lys526Glu mutation. However, a gain-of-function is suggested by general arguments about the pathophysiology of FGFR2 mutations causing Crouzon syndrome.12, 13
The phenotype in this family ranged from mild Crouzon syndrome in the proband to nonpenetrance in her sister. Several other heterozygous mutations of FGFR2 associated with variable phenotypes ranging from mild Crouzon syndrome to clinical nonpenetrance have been described, notably Ser252Leu14 and Ala362Ser.15 A different subset of FGFR2 mutations seems to be associated with nonsyndromic craniosynostosis (without the facial features of Crouzon syndrome) in some individuals and nonpenetrance in others. These mutations include Ala315Ser16 and Ala337Thr (AOMW, SA Wall, unpublished data).
In summary, our new observations emphasise the importance of mutation screening of the FGFR2 tyrosine kinase domain in patients with suspected Crouzon syndrome, who are negative for the usual mutation hotspots (exons IIIa, IIIc), and further explore the blurred boundary between normal and clinical phenotypes in the syndromic craniosynostoses.
References
Cohen Jr MM, Kreiborg S : Birth prevalence studies of the Crouzon syndrome: comparison of direct and indirect methods. Clin Genet 1992; 41: 12–15.
Murdoch-Kinch CA, Ward RE : Metacarpophalangeal analysis in Crouzon syndrome: additional evidence for phenotypic convergence with the acrocephalosyndactyly syndromes. Am J Med Genet 1997; 73: 61–66.
Wilkie AOM, Patey SJ, Kan S-h, van den Ouweland AMW, Hamel BCJ : FGFs, their receptors, and human limb malformations: clinical and molecular correlations. Am J Med Genet 2002; 112: 266–278.
Reardon W, Winter RM, Rutland P, Pulleyn LJ, Jones BM, Malcolm S : Mutations in the fibroblast growth factor receptor 2 gene cause Crouzon syndrome. Nat Genet 1994; 8: 98–103.
Jabs EW, Li X, Scott AF et al: Jackson–Weiss and Crouzon syndromes are allelic with mutations in fibroblast growth factor receptor 2. Nat Genet 1994; 8: 275–279.
Muenke M, Wilkie AOM : Craniosynostosis syndromes; in: Scriver CR, Beaudet AL, Sly WS, Valle D (eds). The Metabolic and Molecular Bases of Inherited Disease, 8th edn. New York: McGraw-Hill, 2000, pp 6117–6146.
Kan S-h, Elanko N, Johnson D et al: Genomic screening of fibroblast growth-factor receptor 2 reveals a wide spectrum of mutations in patients with syndromic craniosynostosis. Am J Hum Genet 2002; 70: 472–486.
Coulier F, Pontarotti P, Roubin R, Hartung H, Goldfarb M, Birnbaum D : Of worms and men: an evolutionary perspective on the fibroblast growth factor (FGF) and FGF receptor families. J Mol Evol 1997; 44: 43–56.
Mohammadi M, Schlessinger J, Hubbard SR : Structure of the FGF receptor tyrosine kinase domain reveals a novel autoinhibitory mechanism. Cell 1996; 86: 577–587.
El Ghouzzi V, Heuertz S, Bonaventure J et al: Mutation analysis in Crouzon and Pfeiffer syndromes identifies novel substitutions in the tyrosine kinase regions of the fibroblast growth factor receptor-2 (FGFR-2). Eur J Hum Genet 2002; 10 (Suppl 1): 247.
Kan SH : Molecular genetics of human limb malformations: HOX genes and FGF pathways; DPhil Thesis, University of Oxford, 2003.
Neilson KM, Friesel RE : Constitutive activation of fibroblast growth factor receptor-2 by a point mutation associated with Crouzon syndrome. J Biol Chem 1995; 270: 26037–26040.
Robertson SC, Meyer AN, Hart KC, Galvin BD, Webster MK, Donoghue DJ : Activating mutations in the extracellular domain of the fibroblast growth factor receptor 2 function by disruption of the disulfide bond in the third immunoglobulin-like domain. Proc Natl Acad Sci USA 1998; 95: 4567–4572.
Oldridge M, Lunt PW, Zackai EH et al: Genotype–phenotype correlation for nucleotide substitutions in the IgII–IgIII linker of FGFR2. Hum Mol Genet 1997; 6: 137–143.
Everett ET, Britto DA, Ward RE, Hartsfield JK : A novel FGFR2 gene mutation in Crouzon syndrome associated with apparent nonpenetrance. Cleft Palate Craniofac J 1999; 36: 533–541.
Johnson D, Wall SA, Mann S, Wilkie AOM : A novel mutation, Ala315Ser, in FGFR2: a gene–environment interaction leading to craniosynostosis? Eur J Hum Genet 2000; 8: 571–577.
Acknowledgements
We thank the family for their help with this study, and the Wellcome Trust for financial support (AOMW).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
de Ravel, T., Taylor, I., Van Oostveldt, A. et al. A further mutation of the FGFR2 tyrosine kinase domain in mild Crouzon syndrome. Eur J Hum Genet 13, 503–505 (2005). https://doi.org/10.1038/sj.ejhg.5201325
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5201325
Keywords
This article is cited by
-
Craniosynostosis as a clinical and diagnostic problem: molecular pathology and genetic counseling
Journal of Applied Genetics (2018)