Abstract
To test the feasibility of developing a diagnostic microarray for a specific disease, we selected all pathogenic changes of the β-globin gene occurring at a frequency ⩾1% in the multi-ethnic Dutch population for analysis. A tagged single-base extension (SBE) approach was used to detect 19 different mutations causing β-thalassemia or abnormal hemoglobins. In the SBE reaction, the primers were elongated at the 3′site with a fluorescently labeled dideoxyribonucleotide triphosphate (ddNTP) complementary to the mutation, following tag hybridization to a glass or flow-through microarray. We compared the performance of a generic glass array and a porous system, by testing each mutation separately using heterozygous carriers and by screening a cohort of 40 unknown β-thalassemia carriers and patients. The results were verified by direct sequencing. The microarray system was able to detect 17 β-globin mutations simultaneously with >95% accuracy in a single SBE reaction. The flow-through array performed slightly better (96%), but the main advantages of the system included real-time data recording and a considerable time saving achieved through a reduced hybridization time.
Similar content being viewed by others
Introduction
The hemoglobinopathies belong to the world's most common recessive monogenic disorders in humans. These involve the α- and β-thalassemias characterized by a deficiency in globin protein synthesis, and the structural mutants of the β-globin chain causing hemoglobin S (HbS), C or E. Worldwide more than 180 different mutations causing β-thalassemia and more than 360 β-globin variants are known.1, 2 Carrier frequencies are particularly high in regions endemic for malaria; however, due to present-day migration patterns, the mutation spectrum in regions without a history of malaria tropica, like in most industrialized North European countries, changed considerably and requires a diagnostic tool, able to combine high-throughput analysis with the capacity to detect a wide spectrum of relatively low frequent mutations.3
In general, carrier detection is performed at the biochemical level with standard hematological methods and confirmation by molecular analysis is usually achieved by direct sequencing. Mutation detection on microarrays is a promising diagnostic tool for high-throughput applications. The feasibility of the technique is often addressed by selecting randomly a number of SNPs associated with disease genes to demonstrate a wide application range.4, 5, 6, 7 Few articles, however, focus on optimization of a larger number of SNPs representing the mutation spectrum for a particular disease gene.8 Here we set out to develop an array for the diagnosis of β-globin defects in populations where many different mutations occur.
Single-base extension
Mutation detection arrays are often based on hybridization of amplified DNA fragments to allele specific oligo (ASO) probes on the array.9, 10 However, these arrays suffer from a high rate of false-positive identifications and are less accurate compared to a minisequence approach.4 Minisequencing is the target-dependent addition by a DNA polymerase of a specific nucleotide to a single primer.11 In an array-based approach, these primer extension reactions are either based on the allele-specificity of the primer, or on extension with a single specific dideoxyribonucleotide triphosphate (ddNTP). In the latter approach, termed single-base extension (SBE), the primer is extended by a DNA polymerase with a single labeled ddNTP that is complementary to the nucleotide at the mutation site. 11, 12, 13, 14 Fan et al15 introduced the tag-SBE, in which the primers contain a unique tail of 20 nucleotides at the 5′ side, complementary to a probe on a generic oligonucleotide array (Figure 1). The tag-probes are independent of the specific mutations to be analyzed, and thus facilitate the choice of probes with similar hybridization efficiencies. This allows flexibility in optimization independent of the array content.
The amplified PCR product is used as a template for the SBE reaction, which is performed in a thermal cycler with a set of 18 mutation-specific primers. Each primer is elongated at the 3′ site with a specific ddNTP, complementary to the wild-type or the mutation. By splitting the SBE reaction into two separate reactions including ddATPCy5/ddUTPCy3/ddGTP/ddCTP and ddGTPCy5/ddCTPCy3/ddATP/ddTTP, all possible combinations can be detected. Each mutation-specific primer contains a unique 20-mer tag at the 5′site that will hybridize to a spotted complementary sequence on the generic array. A fluorescent signal was detected through scanning and a CCD camera for glass and flow-through microarrays, respectively. Open asterisk, Cy3; closed asterisk, Cy5.
Flow-through microarrays
In general, microarrays are spotted on microscope glass slides as a solid support, in which the reaction mixture is added under a cover slide or to a small well created by a silicon overlay.16 Hybridization is dependent on diffusion and environmental factors such as temperature, salt concentration and viscosity, which makes the system inefficient and time consuming. Recently, a flow-through microarray system has been developed that eliminates these limitations by using a porous material as a support for the arrays.17 Target molecules are forced through interconnected capillary pores of an array by flow, resulting in diffusion distances in the order of 100 nm. The hybridization process is therefore fully reaction-rate-limited and complete within a few minutes, allowing real-life detection by CCD camera. Hybridization stringency can be varied by adding/removing solutions and changing the temperature to obtain optimal sensitivity and specificity. In this project we have tested and compared the applicability of flow-through technology for mutation detection to the results obtained using glass arrays.
Materials and methods
Mutation selection and template preparation
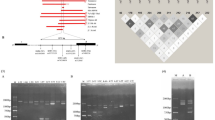
A mutation spectrum for β-thalassemia or β-globin variant defects was determined from 150 patients and carriers living in The Netherlands, diagnosed over a period of 22 months at the Hemoglobinopathies Laboratory, Leiden. In all, 19 mutations occurred with a frequency ⩾1%. These involved five hemoglobin variants (HbS, C, D, E, and J Baltimore) and 14 β-thalassemia mutations, covering 90% of the β-globin mutations in The Netherlands (Figure 2a, Table 1). Iranian DNA samples were obtained from patients treated at the Clinical Center at Bandar Abbas.
(a) Position of the 19 most frequent mutations within the Dutch population in the β-globin gene and the position of the PCR primers. Introns in the β-globin gene are called intervening sequences (IVS). The following primers were used: forward βFF 5′-TGAAGTCCAACTCCTAAGCCA, reverse βER 5′-AAACGATCCTGAGACTTCCA, forward βCF 5′-GTGTACACATATTGACCAAATC and reverse βQ 5′-CCAAGGTTTGAACTAGCTCTTCA. (b) Secondary structure of the IVS2-1 primer. The first 20 5′-nucleotides represent the tag, and the last 23 nucleotides are mutation specific. Five consecutive bases at the 3′ terminus are paired. The folded primer incorporates a labeled ddATP in the SBE reaction. The arrows indicate the site of the base substitution applied to prevent self-annealing of the 3′ terminus.
DNA was extracted from buffy coats isolated from 5–10 ml whole blood by means of a standard salting-out method18 and stored in Tris-HCl EDTA buffer at 4°C. The β-globin gene was PCR amplified in two fragments (Figure 2a)19 using a Robocycler 96 (Stratagene). In all, 45 μl of each PCR product was incubated for 1 h at 37°C with 1.8 U of Shrimp Alkaline Phosphatase (USB) and 18 U of Exonuclease I (USB), inactivated by 15 min at 97°C and purified using QiaQuick columns (Qiagen).
Array preparation and design
The glass coating and spotting protocol was modified from Beier and Hoheisel20 (see www.lgtc.nl). The 20-mer 5′-NH2(C6) modified oligonucleotide probes (Eurogentec) were spotted with a 417™ Arrayer (Affymetrix). Each array contained 19 probes selected from the universal GenFlex® set with a Tm around 58–60°C: control set 000–05, 08, 11, 15, 17, 20, 24, 27, 29, 30, 33, 34, 35, 36, 37, 38, 41, 44, 47 (GenFlex® Tag Array, Affymetrix P/N 610026). Probe 05 was used as a control to determine the hybridization efficiency by the addition of a Cy3- and Cy5-labeled oligo with a complementary tag. Per microscope slide, eight arrays were spotted in triplo. The flow-through arrays were PamChip®'s prepared by PamGene (Den Bosch, NL) as previously described,17 through spotting of 5′-thiolated oligonucleotides (0.2 mm apart) that were covalently coupled to the activated surface. A PamChip® is made of porous Al2O3 with a pore density of 107/mm2, a pore diameter of 200 nm, and a length of 60000 nm, resulting in a surface area of 500 cm2/cm2 and an internal volume of 30 nl/mm2 (for details, visit www.pamgene.com). Each PamChip® contained four sub-arrays with the same 19 tags from the universal GenFlex® set (Affymetrix) spotted in duplicate.
SBE, hybridization and detection
In all, 18 mutation specific primers (20- to 25-mer) for each of the 19 different mutations were designed for both sense and anti-sense SBE reactions (Ta≈61°C). As primer extension reveals the presence of a mutation by incorporation of a differently labeled ddNTP, a single primer can be used to detect both IVS1-1 mutations. Each mutation-specific primer carried a 5′-tag (20-mer) complementary to a single probe spotted on the array. Primer–tag combinations were checked for secondary structure formation with the DNA Mfold program implemented on the bioinfo.math.rpi.edu/∼zukerm site.21
The SBE reaction was performed in a total reaction volume of 30 μl containing 90 ng PCR-template, 18 different SBE primers (0.017 pmol/μl each), two labeled ddNTPs (0.33 pmol/μl each) and two unlabeled ddNTPs (0.66 pmol/μl each; PerkinElmer Life Sciences Inc.), 6.4 U ThermoSequenase (Amersham Biosciences), in 1 × supplied ThermoSequenase buffer. Two separate SBE reactions per DNA sample were carried out – one containing ddATP-Cy3, ddUTP-Cy5, ddCTP and ddGTP, and another containing ddATP, ddTTP, ddCTP-Cy3 and ddGTP-Cy5 – to allow the detection of all possible mutations using a two- channel scanner device (418™ Scanner, Affymetrix, filters 532 and 635 nm for Cy3 and Cy5 detection, respectively). The SBE reaction was performed with an initial denaturation step 3.5 min at 92°C and 40 two-step cycles of 45 s 92°C and 30 s 61°C in Robocycler 96 (Stratagene).
The glass slide was covered with a punched rubber overlay forming eight incubation chambers, one triplo array per incubation chamber, put into a home-made Perspex holder and cover with eight holes for the addition of sample, kept in place by clamps. In all, 10 μl of the SBE reaction was added to 50 μl of hybridization solution (3.4 × SSC, 0.3% SDS (w/v) in Tris EDTA) and added to a well on the glass array. During hybridization at 40°C o.n., the holes were sealed with tape to prevent evaporation. The arrays were washed for 5 min subsequently in 2 × SSC, 70% ethanol and 100% ethanol, and air-dried.
For the PamChip®, incubation and detection was performed on a prototype of the FD10 Pam Microarray system (PamGene and Olympus Optical). Incubations were performed in a thermostatically controlled incubator holding one chip, containing four arrays. Prior to hybridization, each array was washed at 40°C with respectively 20 μl of PBS/0.1% (w/v) Tween 20 and 20 μl of 3 × SSP (20 × SSP=180 mmol/l NaCl and 10 mmol/l Sodium phosphate). Both solutions were pumped one cycle (30 s) down and up through the pores of the array using a Microlab 500 syringe pump (Hamilton). A measure of 10 μl of the SBE reaction was diluted with 7 μl of ddH2O, boiled for 5 min and cooled on ice. After addition of 3 μl of 20 × SSP, the solution was hybridized for 5 min at 40°C. The solution was pumped down, and images were captured with an 8-bit CCD camera.
All arrays were analyzed using ArrayPro (Media Cybernetics). The spot intensity was calculated after reduction of the median spot intensity with the median local background intensity. The signal was scored positive when it was found at least twice in the same array with an intensity above 5 on a scale of 0–255.
Results
SBE optimization
In all, 18 sense and anti-sense primers were designed to detect 19 mutations and tested individually on a wild-type template. Nonspecific or no signal was observed in seven sense and eight anti-sense primers. We have chosen to optimize the sense primers, except for the anti-sense Cd121, which performed as required (Table 2). For the primers used to detect the Cd16, IVS2-1 and Cd39 mutations, a false-positive signal was observed even in the absence of the template. Analysis in DNA Mfold21 predicted the formation of a stable stem loop with three to five consecutive base pairings at the 3′ end (Figure 2b), which could explain the auto-extension signal during SBE. New primers were designed containing an internal mutation to prevent self-annealing at the 3′ terminus for Cd16, IVS2-1 and Cd39. The modified primers showed an increase in specificity by the loss of auto-extension, but the signal for Cd39 dropped below the detection level. The original Cd39 primer could still be used in the assay, because the false-positive signal with ddATP did not interfere with the capacity of the primer to distinguish between the wild-type (ddCTP) and mutant (ddTTP) allele.
The effect of primer concentration on signal intensity was tested by using seven different dilutions in a multiplex of four primers, ranging from 5 to 0.125 pmol in a 30 μl SBE reaction (Figure 3a). The signal for primers Cd39, Cd41-42 and IVS1-1 was optimal at 0.5 pmol each, and no optimum was seen for the Cd121 primer. In the final multiplex SBE containing 18 mutation-specific primers, a concentration of 0.5 pmol/30 μl for each primer was used.
(a) Wild-type signal intensity found with four different primers in seven different concentrations. The SBE reaction was performed on wild-type PCR product and hybridized to a glass array. (b) Scans of glass and flow-through arrays and the corresponding signal after hybridization of a compound heterozygous patient HbS(A → T)/IVS1-110(G → A). Each array contains four rows and six columns. Marker spots are located on the glass array at the first and fourth row, first and fourth columns. The spots for HbS and IVS1-110 are indicated by arrows. The positions of the probes are different for the glass and flow-through array: on the left, the scans of the glass array for each nucleotide (first row, fifth column HbS spot; third row, fourth column IVS1-110 spot), and on the right the flow-through array (third row, fourth column HbS spot; fourth row, third column IVS1-110 spot). Images of glass arrays were taken after overnight hybridization and subsequent wash; images of the flow-through arrays were taken after 5 min hybridization with subsequent pumping down of the fluid and no wash. In the graphs below, the signal for the HbS and IVS1-110 spot for each possible extension with a ddNTP is given.
The ability of the system to detect all of the 19 mutations simultaneously in a single SBE reaction was tested. Out of 19 heterozygous carriers, 17 were scored correctly, that is, both the wild-type and the mutant signal were detectable. No mutant signal was observed for Cd8 and Cd8/9, and these primers were omitted from further analysis. For all mutations, we observed a reduction in wild-type signal to half the intensity in the heterozygotes as compared to the homozygous wild-types.
Screening for β-globin mutations
The specificity of the system was tested in a blinded screening of 20 selected Dutch and 20 Iranian samples (n=80 alleles) for which the genotypes were confirmed by direct sequencing. These include 23 wild-type and 57 mutant alleles, representing 14 different mutations. Per sample a single SBE reaction with the 17 SBE primers was performed, one-third was used for hybridization to the glass-array and one-third for the flow-through (Figure 3b). Out of 80 alleles, 76 alleles were scored correctly on glass (95%) and 77 on PamChip® (96%). A homozygote HbE was once incorrectly scored as a heterozygous on the glass array, while on the flow-through array a heterozygous carrier for HbE was scored as a homozygote. Both on glass and flow-through array a heterozygote for Cd41/42 was scored as wild-type due to low signal, and the compound heterozygote Cd41-42/IVS1-5 was erroneously scored as a combination of homozygosity for mutation Cd41-42 and heterozygosity for IVS1-5. The Cd39 mutation in the compound heterozygote Cd39/IVS2-1 was missed on the glass array.
Discussion
A microarray system, using tagged SBE and hybridization to generic home-made glass arrays and commercial PamChip®, was developed to detect 19 different β-thalassemia and β-globin variant mutations simultaneously. The SBE reaction and hybridization conditions to the array were optimized using a wild-type template and the specificity was tested on PCR templates heterozygous for each mutation. Out of 19 mutations, 17 could be detected reproducibly, which cover more than 80% of the mutation spectrum observed in The Netherlands and other industrialized North European countries with similar mutation spectra due to recent immigration. The specificity of the system was tested on 40 sequenced carriers and patients samples. Out of 80 alleles, the genotype was correctly assigned in 76 using the glass array and 77 with PamChip®, reaching a 95% reliability. As the selected set includes the most frequent Mediterranean, Asian and African mutations, the system is widely applicable for high-throughput diagnostic purposes in many different countries.
Factors that influence signal intensity and specificity are primer concentration, hybridization efficiency and primer sequence. The primer concentration was varied to reach an optimal signal intensity. If the concentration is too low, the signal remains below detection level, while higher amounts are not extended efficiently, causing competition between tags of extended and nonextended primers for the probes on the array resulting in an overall reduction in signal.
In three mutation-specific tag–primers (cd16, IVS2-1 and cd39), a false-positive signal was observed in the absence of template. Template-independent primer extension might be due to the formation of secondary structures. This has occasionally been observed, particularly in assay formats based on immobilized primers.4, 14 According to Pastinen et al,4 this problem can be avoided by performing the minisequencing reactions at an elevated temperature or by analyzing the other strand of the DNA template. In our study, SBE was performed at a relatively high temperature of 61°C, which did not prevent the problem. We were able to avoid the false signal in two cases by altering the primer at the base complementary to the 3′ terminus (Figure 2b).
Out of the 19 mutations tested, three are frameshift mutations involving insertion or deletion of one or several nucleotides. Remarkably, two of these, that is, Cd8(−AA) and cd8/9(+G), did not show the correct genotype when tested in homo- or heterozygotes, while the cd41-42(−TCTT) was among the alleles scored erroneously in the blind screening of 80 alleles, due to low mutant signal intensity. It is not clear however, how this relates to secondary structures in the PCR products. Apparently, incorporation efficiencies seem to be largely dependent on the local sequence environment. Syvänen5 suggested that the labels might affect the specificity of the nucleotide incorporation by the DNA polymerase. However, overall preference for incorporation of a specific labeled nucleotide was not observed among the 17 other primers included in the set.
Other techniques frequently used to detect β-globin mutations in a diagnostic setting are ASO probe hybridization,22 denaturing gel gradient electrophoresis (DGGE),19 and amplification refractory mutation system (ARMS).23 Most of these approaches are time consuming, require multiple reactions per patient, radioactive reagents or expensive laboratory equipment, and are not always suited for high-throughput analysis. The SBE and hybridization to generic arrays show high specificity (95%) and flexibility, since no new arrays are needed to change the set of mutations to be analyzed, making the system easily adaptable to changing reasearch goals. As glass coating is performed on microscope slides with relatively simple chemicals and only minimal amounts of probes are used by the 417™ arrayer, arrays can be produced at low cost (€0.12 per array). The total price for the SBE reaction is approximately €8;- per sample, the majority of the expenses involve the enzymes and labeled ddNTPs. This is competitive with direct sequencing of the complete β-globin gene, as presently used by many diagnostic laboratories.
Our experiments clearly show that the probes can be easily transferred from the glass to the PamChip® microarray system. The main advantage of the flow-through system is the considerably reduced hybridization time, which lasts only 5 min compared to 16 h for glass arrays. Additional time is saved by sample preparation and by the real-time recording of hybridization results by the CCD camera, which permits an automated registration and analysis of data. The ability of the system to vary hybridization stringency allows temperature-dependent optimization of mutations, not detectable under normal circumstances. These possibilities will be explored in future studies.
References
Weatherall DJ, Clegg JB : The Thalassaemia Syndromes. Oxford: Blackwell Publishing; 1981, (3rd edn).
Thein SL : Beta-thalassaemia. Baillières Clin. Haematol. 1998; 11: 91–126.
Giordano PC, Harteveld CL, Heister AJGM et al: The molecular spectrum of beta-thalassemia and abnormal hemoglobins in the allochtonous and autochtonous Dutch population. Community Genet 1998; 1: 243–251.
Pastinen T, Kurg A, Metspalu A, Peltonen L, Syvänen AC : Minisequencing: a specific tool for DNA analysis and diagnostics on oligonucleotide arrays. Genome Res 1997; 7: 606–614.
Syvänen AC : From gels to chips: ‘Minisequencing’ primer extension for analysis of point mutations and single nucleotide polymorphisms. 1999 [Review]. Hum Mutat 1999; 13: 1–10.
Gerry NP, Witowski NE, Day J, Hammer RP, Barany G, Barany F : Universal DNA microarray method for multiplex detection of low abundance point mutations. J Mol Biol 1999; 292: 251–262.
Fortina P, Delgrosse K, Sakamuze T et al: Simple two-color array-based approach for mutation detection. Eur J Hum Genet 2000; 8: 884–894.
Gemignani F, Perra C, Landi S et al: Reliable Detection of β-Thalassemia and G6PD Mutations by a DNA Microarray. Clin Chem 2002; 48: 2051–2054.
Hacia JG, Brody LC, Chee MS, Fodor SP, Collins FS : Detection of heterozygous mutations in BRCA1 using high density oligonucleotide arrays and two-colour fluorescence analysis. Nat Genet 1996; 14: 441–447.
Wang DG, Fan JB, Siao CJ et al: Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science 1998; 15 280: 1077–1082.
Syvänen AC, Aalto-Setälä K, Harju L, Kontula K, Söderlund H : A primer-guided nucleotide incorporation assay in the genotyping of apolipoprotein E. Genomics 1990; 8: 684–692.
Sokolov BP : Primer extension technique for the detection of single nucleotides in genomic DNA. Nucleic Acids Res 1990; 18: 3671.
Kuppuswami MN, Hoffmann JW, Kasper CK, Spitzer SG, Groce SL : Single-nucleotide primer extension to detect genetic diseases: experimental application to hemophilia B (factor IX) and cystic fibrosis genes. Proc Natl Acad Sci USA 1991; 88: 1143–1147.
Nikiforov TT, Rendle RB, Goelet P et al.: Genetic bit analysis: a solid-phase method for typing single nucleotide polymorphisms. Nucleic Acids Res 1994; 22: 4167–4175.
Fan J-B, Chen X, Halushka MK et al: Parallel genotyping of human SNPs using generic high-density oligonucleotide tag arrays. Genome Res 2000; 10: 853–860.
Pastinen T, Raitio M, Lindroos K, Tainola P, Peltonen L, Syvänen AC : A system for specific, high-throughput genotyping by allele-specific primer extension on microarrays. Genome Res 2000; 10: 1031–1042.
Beuningen R van, van Damme H, Boender P, Bastiaensen N, Chan A, Kievits T : Fast and specific hybridization using flow-through microarrays on porous metal oxide. Clin Chem 2001; 47: 1931–1933.
Miller SA, Dykes DD, Polesky HF : A simple salting-out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988; 16: 1215.
Losekoot M, Fodde R, Harteveld CL, van Heeren H, Giordano PC, Bernini LF : Denaturing gradient gel electrophoresis and direct sequencing of PCR amplified genomic DNA: a rapid and reliable diagnostic approach to beta-thalassaemia. Br J Haematol 1990; 76: 269–274.
Beier M, Hoheisel JD : Versatile derivatisation of solid support media for covalent bonding on DNA-microchips. Nucleic Acids Res 1999; 27: 1970–1977.
SantaLucia Jr J : A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc Natl Acad Sci USA. 1998; 95: 1460–1465.
Cai SP, Wall J, Kan YW, Chehab FF : Reverse dot blot probes fot the screening of β-thalassemia mutations in Asians and American Blacks. Hum Mutat 1994; 3: 59–63.
Old JM, Varawella NY, Weatherall DJ : Rapid detection and prenatal diagnosis of β-thalassemia: studies in Indian and Cypriot populations in the UK. Lancet 1990; 336: 834–837.
Acknowledgements
The authors thank Victor de Jager for optimization of the glass-coating protocol and Majid Yavarian for providing the Iranian samples.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
van Moorsel, C., van Wijngaarden, E., Fokkema, I. et al. β-Globin mutation detection by tagged single-base extension and hybridization to universal glass and flow-through microarrays. Eur J Hum Genet 12, 567–573 (2004). https://doi.org/10.1038/sj.ejhg.5201192
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5201192
Keywords
This article is cited by
-
Development of a fibrous DNA chip for cost-effective β-thalassemia genotyping
International Journal of Hematology (2012)
-
Development of a Flow-Trough Microarray based Reverse Transcriptase Multiplex Ligation-Dependent Probe Amplification Assay for the Detection of European Bunyaviruses
Molecular Biotechnology (2011)
-
TAMGeS: a Three-Array Method for Genotyping of SNPs by a dual-colour approach
BMC Genomics (2007)
-
Microarray MAPH: accurate array-based detection of relative copy number in genomic DNA
BMC Genomics (2006)