Abstract
We used fluorescent in situ hybridisation (FISH) to detect nine chromosomes (1, 13, 15, 16, 17, 18, 21, 22 and X) in 89 first Polar Bodies (1PBs), from in vitro matured oocytes discarded from IVF cycles. In 54 1PBs, we also analysed the corresponding oocyte in metaphase II (MII) to confirm the results; the other 35 1PBs were analysed alone as when preimplantation genetic diagnosis using 1PB (PGD-1PB) is performed. The frequency of aneuploid oocytes found was 47.5%; if the risk of aneuploidy for 23 chromosomes is estimated, the percentage rises to 57.2%. Missing chromosomes or chromatids found in 1PBs of 1PB/MII doublets were confirmed by MII results in 74.2%, indicating that only 25.8% of them were artefactual. Abnormalities observed in 1PBs were 55.8% whole-chromosome alterations and 44.2% chromatid anomalies. We observed a balanced predivision of chromatids for all chromosomes analysed. Differences between balanced predivision in 1PB and MII were statistically significant (P<0.0001, χ2 test); the 1PB was most affected. The mean abnormal segregation frequency for each chromosome was 0.89% (range 0.52–1.70%); so, each of the 23 chromosomes of an oocyte has a risk of 0.89% to be involved in aneuploidy. No significant differences were observed regarding age, type of abnormality (chromosome or chromatid alterations) or frequency of aneuploidy. Nine of the 35 patients (25.7%) whose 1PB and MII were studied presented abnormalities (extra chromosomes) that probably originated in early oogenesis. Analysis of 1PBs to select euploid oocytes could help patients of advanced age undergoing in vitro fertilization (IVF) treatment.
Similar content being viewed by others
Introduction
Fertility decreases with advanced maternal age, both in fertile and infertile couples. In general, the probability to obtain a pregnancy per cycle is 25%, but most people who attend assisted reproduction centres have fertility rates of less than 10% and assisted reproduction techniques (ART) improve their chances. Younger patients have better fertility rates after ART. Approximately, 30% of the 35 to 39-year-old women have fertility problems and, consequently, the average age of ART patients is high. It is important to study the reasons why a decrease in the success of ART occurs at these ages.
The ability of the embryo to implant appears to be the most important factor for the decline in fertility with advanced maternal age. Some endometrial factors could also contribute to the increasing difficulty to establish a pregnancy.1 Another cause of this decrease is poor embryo quality that, in part, results from a poor oocyte quality.2 Embryo aneuploidy is another possible reason. Aneuploidy has been found to increase significantly with maternal age in several studies.3,4,5,6,7 Frequencies of 4.9, 11.5 and 29.8% aneuploid oocytes in the 25–34, 35–39 and 40–45 maternal age groups have been estimated.8
At the eight-cell stage, an increase of X, Y, 13, 18 and 21 aneuploidy from 4% in women 25–34 years old to 37% in women aged 40 years or more has been found using fluorescent in-situ hybridisation (FISH)9; a similar result has been obtained in oocytes.8,10,11,12
Methods to select chromosomically normal embryos based on morphology13 or capacity to grow to the blastocyst stage14 have been sought, but these approaches have failed. Between 15 and 66% morphologically normal embryos are aneuploid9,15,16 and 37% of trisomic embryos reach the blastocyst stage.17
The influence of the most common chromosome abnormalities at birth (X, Y, 13, 18 and 21) and of those that could increase the abortion rate and decrease implantation chances in these patients have been analysed through preimplantation genetic diagnosis (PGD) in blastomeres by FISH. In some studies up to nine different chromosomes are analysed18,19,20 and the success of ART is slightly improved. A reduction in abortions19,20 and even an increase in implantation rate20 has been observed, although the percentage of pregnancies has not increased.21
Since, as reported, 90% of embryo aneuploidy is the result of nondisjunction in maternal meiosis I,22 detection of abnormal oocytes through the analysis of their first polar body (1PB) in an in vitro fertilisation (IVF) treatment is an important goal.
In meiosis I, one set of chromosomes in the oocyte (with two chromatids per chromosome) segregates to the 1PB and the oocyte in metaphase II (MII) retains a chromosome constitution which is the mirror image of the one found in the 1PB. This allows the indirect characterisation of the chromosome constitution of the oocyte through the study of the 1PB.23 By analysing 1PBs, errors related to embryo mosaicism can be avoided, there is time to perform up to three rounds of FISH and, if problems have occurred during the process, a blastomere can be biopsied and analysed on time before transfer. Recently, a preimplantation genetic diagnosis using IPB (PB (PGD−1PB)) protocol in which ICSI was performed before 1PB biopsy was applied,24 and the fertilisation rate achieved was comparable to the one obtained in IVF-ICSI cycles without PGD. Moreover, different types of FISH probes, (CEP), centromeric locus specific (LSI) and whole chromosome painting (WCP) can be used because the 1PB is in metaphase, and, when fixed, chromosome spreads are obtained. This technique has been applied when the female is a carrier of structural chromosome abnormalities (reviewed in Durban et al.24) or in patients with a high risk of aneuploidy. In the latter case, up to five different chromosomes have been analysed.25,26,27. The detection of abnormal oocytes through the analysis of their 1PB could help to lower the abortion rate and to increase implantation chances, and both effects should increase the incidence of pregnancy in some patients, including those of advanced maternal age.
Here, we describe our results in the detection of aneuploidies in mature oocytes. Initially, discarded oocytes from IVF cycles, most of them matured in vitro, were used. We analysed the incidence of the most common autosomal chromosome abnormalities25,26,28 as well as aneuploidies for chromosomes X, 1, 15 and 17 which could also contribute to the low pregnancy rates mentioned.29
Then, we optimised the analysis of nine chromosomes in 1PB extensions, and to evaluate its feasibility, we compared the results obtained in fixed 1PB with those obtained in their corresponding oocytes in MII. An evaluation of hypohaploidies was made in this way. We also evaluated aneuploidy rates in different age groups to confirm the age where screening of aneuploidies would be appropriate. Third, we analysed which chromosomes were more implicated in aneuploidies and in which ones the interpretation of the FISH signals was more difficult and, consequently, the diagnosis would have to be more accurate.
Materials and methods
A total of 89 oocytes donated by 60 women 18–45 years old (mean 33.7) who, due to different aetiologies, were included in an IVF programme (Institut Universitari Dexeus, Barcelona) were analysed. Written informed consent was obtained and the project was approved by our institutional ethics committee. Patients underwent gonadotropin stimulation.30 Dosages were adjusted based on their response to treatment. Oocytes were retrieved by transvaginal follicular puncture under ultrasound scanning control.
The cells used were as follows (Figure 1): three oocytes matured in vivo donated by one patient; 78 oocytes matured in vitro that were cultured 24–48 h because they were at the germinal vesicle (GV) or metaphase I stage the day of follicular puncture (D+0), 68 of them were fixed the day after oocyte retrieval (D+1) and 10 were fixed 2 days after oocyte retrieval (D+2); and eight unfertilised oocytes inseminated by conventional IVF, but in which no pronuclei were observed, they were fixed on D+1.
We fixed and analysed the 1PB of all 89 oocytes, and in 54 of them we also fixed and analysed the corresponding MII. Since the technique was developed to be applied in PGD cycles using 1PBs, most 1PBs were fixed separately from the oocyte in MII as in a diagnostic procedure, and only 16 were fixed together with the egg. To separate the 1PB from the oocyte in metaphase MII, a regular 1PB biopsy was used in 73 oocytes. To biopsy 1PBs, oocytes were placed in 50 μl buffered medium drops (Gamete, Vitrolife, Göteborg) covered by mineral oil (Ovoil, Vitrolife). A 20 μm diameter hole was made in the zona pelucida (ZP) of the oocytes using a laser beam (1.48 μm noncontact diode laser; Fertilase, Medical Technologies, Montreux, Switzerland)31 in 69 oocytes or a mechanical method (PZD) in four cases. To remove the 1PB, hand-made and commercial micropipettes (Cook, Australia) of 25–30 μm external diameter were used.
Fixation of 1PBs was made using a modification of Durban et al,24,32 which helps to reduce chromosome losses to a minimum and practically eliminates artefactual errors. Oocytes were fixed using a modification of a described method.33 Fixed cells were observed in a phase contrast microscope (Olympus BX 60) and captured and saved in a computer Power Macintosh G3 equipped with Quips Lab Manager software (Vysis, NY, USA). To eliminate cytoplasmic debris, in some of them, a 0.5–3 min enzymatic treatment using pepsin (Sigma, Spain) was applied (50 μg/ml in HCl 10 mM at 37°C, rinsing at room temperature with purified water and allowed to air-dry) before adding a drop of freshly prepared Carnoy fixative (methanol:acetic acid, 3:1). Most slides were frozen at −20°C until they were hybridised.
Prior to FISH, preparations were treated with 50 mM MgCl2/PBS (4 min), postfixed with 3% formaldehyde in 50 mM MgCl2/ PBS (8 min), cleansed in PBS (4 min) and dehydrated in 70, 80 and 95% ethanol (2 min each). They were stained with 4,6-diamidino-2-phenylindole (25% DAPI II, Vysis, NY, USA / 75% Antifade, Vectashield, CA, USA) and observed in a fluorescence microscope. DAPI images were also saved to compare them with phase contrast and FISH images.
DAPI was eliminated by rinsing slides with 0.1% Tween 20/2 × SSC (2 × 5 min) at room under agitation, dehydrated in 70, 80 and 95% ethanol (4 min each), air-dried and then dried in a hot plate at 45°C for 4 min. Two rounds of FISH were performed with several fluorescent probe combinations. In the first round, chromosomes 13, 16, 18, 21 and 22 labelled, respectively, with SpectrumRed, SpectrumAqua, SpectrumBlue, SpectrumGreen and SpectrumGold (MultiVysionPB, Vysis, NY, USA) were analysed. In the second round, a mixture of X, 1, 15 and 17 probes labelled, respectively, with SpectrumAqua, SpectrumOrange, SpectrumGreen and SpectrumOrange/SpectrumGreen (Vysis, NY, USA) was applied.
The hybridisation solution was applied (0.5 μl) to each slide containing one (1PB or MII) or two (1PB and MII) fixed cells, and covered with a 6 × 6 mm coverslip. The slides were sealed with rubber cement and placed for 5 min on a hot-plate at 72°C to co-denature nuclear DNA with the probes; they were then allowed to hybridise overnight at 37°C in a dark moist chamber. In our hands, good results in the second round of FISH could also be obtained using a microwave during 45 min at 75 W instead of an overnight incubation. After each hybridisation round, slides were washed using the formamide or the rapid wash procedures recommended by the probe manufacturer. Slides were mounted with 8 μl DAPI II (Vysis, NY, USA) or, in the case of the MultiVysion probe, with 8 μl Antifade (Vectashield, CA, USA). Visualisation was made under an Olympus BX 60 microscope equipped with a high-sensitivity camera (Roger Scientific, Photometrics, Tucson, Arizona USA) and with filters for the fluorochromes used and connected to a computer Power Macintosh G3 with software for Smartcapture (Digital Scientific Cambridge, UK) and IPLab (Scanalytics, Inc., Vysis, USA) which allowed the analysis.
After visualisation and analysis of every set of probes and before the following round of FISH, slides were washed as previously described.
Scoring criteria for aneuploidies
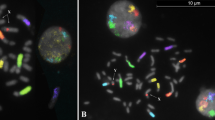
As a result of the possible chromosome loss while spreading the cells on the slide or of FISH failure, we considered as confirmed aneuploidies for the chromosomes analysed the presence of additional signals. Missing chromosomes or chromatids were only considered when the result was confirmed in the complementary MII (Figure 2, parts 4b and 4d).
1PBs (a and b) and their corresponding oocytes in MII (c and d). The colours are the same as the probes employed. Arrows indicate abnormalities. 1: DAPI stained 1PB and the corresponding MII. The different chromosome packaging of the two cells can be seen. 2: 1PB and the corresponding MII. Both cells were euploid for the chromosomes analysed. 3: 1PB and the corresponding MII with multiple aneuploidies. 1PB with missing chromosomes 1, 15, 17 and 22 and chromatids of chromosomes 13 and 18. MII with extra chromosomes 1, 15, 17 and 22 and chromatids of chromosomes 13 and 18. 4: Second round of FISH showing a 1PB with a missing chromosome 1 and X and the corresponding MII with and extra chromosome 1 and X. The first round showed euploidy. 5: Second round of FISH showing a 1PB with an extra chromatid of chromosome X and the corresponding euploid MII. The first round showed euploidy.
We considered balanced chromatid predivision to occur when the signals for the two chromatids were separated by more than the distance that would allow for two additional signals.8
Results
In this work, from a total of 89 mature oocytes, we analysed 54 1PB/MII doublets and 35 single 1PBs (Table 1).
As seen in Figure 2, part 1, fuzzy 1PB chromosomes can be distinguished from MII chromosomes. When LSI or CEP probes are used, two spots per chromosome are usually visualised, corresponding each to a chromatid.
Out of 89 1PBs, 73 could be diagnosed (82%) while 16 were not (18%). Of the diagnosed 1PBs, 33 were euploid (Figure 2, parts 2a–2d), 17 hyperhaploid and 23 hypohaploid. Most aneuploid 1PBs had one (26) or two (10) chromosome or chromatid abnormalities, while in four cells three (2), four (1) or six chromosomes (1) were affected (Figure 2, parts 3a–3d). These values fit a Poisson with a parameter of 0.849 (calculated according to Kolmogorov–Smirnov adjustment). All cytogenetic alterations indicated are listed in Table 2.
In the 1PBs in which the complementary MII was analysed, 23 out of 31 (74.2%) missing chromosomes or chromatids in the 1PB were found in the MII. We extrapolated this percentage to the missing chromosomes or chromatids found in 1PBs analysed alone. We considered that each missing chromosome or chromatid found in a single 1PB contributed 0.74 instead of 1 when counting the number of abnormal chromosomes (Table 3). In the 89 analysed 1PBs, we found a total of 57.6 altered chromosomes (13.6 belonging to single 1PBs and 44 in 1PBs that had the complementary MII). Looking at the frequency of alteration found for each of the nine analysed chromosomes, chromosome 16 showed the highest rate followed by chromosomes 13, 22, 17, 21, X, 15, 1 and 18 (Table 3). The frequencies were not significantly different from the expected rate (χ2 test). No relation was found between the chromosomes implicated in aneuploidies and their size (classifying the chromosomes as long, medium-sized and small) or morphology (classifying them as acrocentrics and nonacrocentrics) when applying a χ2 test.
For the chromosomes analysed, the abnormal segregation percentage of each chromosome varied between 0.52 and 1.70%, with an average of 0.89% (data calculated from Table 3). According to these results, each of the 23 chromosomes of an oocyte has a 0.89% risk of being involved in aneuploidy.
The abnormalities observed in 1PBs consisted of 55.8% whole-chromosome alterations and 44.2% chromatid anomalies. The number of chromosome or chromatid errors was very similar for chromosomes X, 13, 16, 17 and 22 but not for chromosomes 1, 15, 18 and 21.
We observed a balanced predivision of chromatids in all chromosomes analysed (Table 4). The difference between balanced predivision in 1PB and MII was statistically significant (P<0.0001, χ2 test), with the 1PB most affected. In chromosomes 1, 13, 15, 17 and X a significant difference was observed between the balanced predivision frequency found in 1PB and in MII. In 1PB, the frequencies per individual chromosomes ranged from 12.5 to 20.9%, although significant differences were not observed.
Global aneuploidy rate
In the group of 1PBs analysed with the corresponding oocyte in MII, seven were found to be hypohaploid; in all cases, a single chromosome was affected; six of the seven hypohaploidies were confirmed by the presence of the missing chromosome in the MII. This means that in one out of seven hypohaploid 1PBs the result was not confirmed (14.2%). In the group of 1PBs analysed alone, nine were found to be hypohaploid for a single chromosome. We considered that 1.3 (9 × 0.142) of this hypohaploid 1PBs would not be confirmed if the MII had been analysed. According to this, we counted as aneuploid cells eight hypohaploid single 1PBs instead of nine (Table 5).
After diagnoses of the chromosomes in the 1PB or in the MII (Table 5), instead of only considering 1PB results, 100% of diagnosis was achieved in 1PB/MII (54/54) doublets (the oocyte with a nonconfirmed missing chromosome in the 1PB was considered euploid, as shown by the result in the corresponding MII). A total of 28 oocytes were euploid, 13 were hyperhaploid and 13 were hypohaploid. Adding also 1PBs analysed alone, we obtained a total of 80 out of 89 diagnosed cells (90%). In all, 42 were euploid, 17 hyperhaploid and 21 hypohaploid. The final frequency of aneuploid oocytes found in the present study was 47.5% (38/80).
Relation between aneuploidy and age
We found aneuploid 1PBs in 32 women, 14 of them ≥35 years old and 18<35 years old (Table 6). A total of 39 1PBs from these women, ranging in age from 28 to 45 years (mean 34.5), were aneuploid. In total, 17 aneuploid cells were from women ≥35 years (mean 37.5 years) and 22 from women <35 years old (mean 31.6). It is worth to emphasising that in a 31-year-old patient (patient S), we found four aneuploid cells from a total of five analysed 1PBs. Patient AE had two aneuploid 1PBs out of four and was a Rob(13;14) carrier; the aneuploidies affected chromosomes not involved in the rearrangement.
We considered as aneuploid 1PBs those that were hyperhaploid and those confirmed as hypohaploid in 1PB/MII doublets, as well as all aneuploid 1PBs analysed alone to avoid underestimating the aneuploidy rate for each chromosome, because by using the corrected evaluation of aneuploidy, one single 1PB should have been eliminated at random.
An ANOVA and a Student's t-test showed no significant differences when considering age and either type of abnormality (chromosome or chromatid alterations) or the frequency of aneuploidy observed.
Presence of extra chromosomes
A total of 11 1PB/MII doublets had additional chromosomes (Table 7). An extra copy of a chromosome or chromatid was seen in one cell while the other had the normal chromosome (Figures 2, 5b and 5d). Moreover, one oocyte with an extra chromosome in the 1PB was not included in this group because the MII was undiagnosed; this cell had a missing chromatid 13. Nine of the 35 patients (25.7%) whose 1PB and MII were studied had oocytes with extra chromosomes. The mean age of these patients was 34 years (ranging from 31 to 38). Five patients were <35 years and four patients were ≥35 years. Patient S, who was only 31 years old, had three of the 11 mentioned cells.
Discussion
In this study, we analysed 35 single 1PBs, and through the analysis of 54 1PB/MII doublets, we could estimate the percentage of missing chromosomes or chromatids that would be artefactual when analysing single 1PBs (26%). According to our results, a hypohaploidy would have 74% probability of resulting from an aneuploidy event, that is, in our hands, only one-fourth of hypohaploidies would be artefactual.
In the second round of FISH, results were better when using a microwave34,35 instead of an overnight incubation at 37°C. Probably, the agitation of the water molecules induced by the microwaves helped to improve the homologous recognition between the nuclear DNA and the probes. In our opinion, its use in PGD cycles could help minimise the time to provide a diagnosis.
We could diagnose 80 out of 89 analysed oocytes (90%), although when considering only 1PB results we diagnosed 73 out of 89 (82%) cells. In practice, when one or more chromosome anomalies have been detected, hybridisation failures make no difference because the cell is considered aneuploid. On the other hand, in those cells that are euploid for the chromosomes diagnosed but in which some chromosomes are undiagnosed, the problem becomes evident, mainly when undiagnosed chromosomes could produce viable aneuploid embryos. In our case, this happened in seven 1PBs.
All nine chromosomes were diagnosed in 60 1PBs (67.4%), while in the other 29 1PBs (32.6%) one or more chromosomes were undiagnosed: in 20 of them (22.5%) seven or eight chromosomes were diagnosed, while in nine 1PBs (10.1%) less than seven chromosomes could be diagnosed. The misdiagnoses were either because the cell was not found by FISH or because of the difficulty of assigning diffuse signals to a chromosome or a chromatid. We found this difficulty in all analysed chromosomes independent of whether the probe used was CEP or LSI. LSI 22 was the most affected and CEP 18 the least (Table 3).
The diagnostic efficiency of each of the chromosomes analysed in the first or in the second round of FISH was not different; however, we found a higher frequency of nondiagnosis in the second round. This was because some cells (four) were lost during the application of the second FISH panel.
Chromosomes more implicated in aneuploidy
The highest rate of aneuploidy was found for chromosome 16 (1.70%), as might be expected, followed by chromosome 13 (1.35%), although the frequencies were not significantly different from the expected rate (χ2 test). Trisomy 16 is the most frequent trisomy in spontaneous abortion studies.36
On the other hand, chromosome 21 had the highest rate of aneuploidy in unfertilised oocytes;37 however, in a study of fresh oocytes using the SKY Vysion spectral imaging system,38 chromosomes 14 and 22 were the most implicated in aneuploidies.
As mentioned, we found aneuploidies for all chromosomes analysed but no relation was found between their frequency and chromosome size or morphology (χ2 test). In contrast with our results, other authors did not find aneuploidies for chromosomes 1 and 9.37
Estimation of the risk for each chromosome of being implicated in aneuploidy
Extra and missing chromosomes and chromatids were summed up and divided by the total number of diagnosed chromosomes either in single 1PBs or in 1PBs with the MII analysed to estimate the risk of aneuploidy for each chromosome analysed. We observed (data calculated from Table 3) that for the chromosomes analysed, the abnormal segregation percentage for each chromosome ranged from 0.52 to 1.70%, with an average of 0.89%.
In the present study, analysing nine chromosomes, a 47.5% aneuploidy rate was found. Probably, a higher aneuploidy rate would be found if more chromosomes had been analysed. Since the incidence of oocytes with aneuploidies fits a Poisson with a 0.849 parameter, we were able to make an estimation of this aneuploidy rate. According to the frequency values of this distribution, the probability of having a euploid oocyte would be 42.8%. The other 57.2% of cells would be aneuploid, and 36.3, 15.4, 4.4, 0.9 and 0.2% would be aneuploid for one, two, three, four and five chromosomes, respectively.
Chromosome and chromatid alterations
The abnormalities observed in 1PBs and oocytes included 55.8% whole-chromosome alterations and 44.2% chromatid anomalies. Some authors suggested a similar distribution of abnormalities derived from chromosomal non-disjunctions and chromatid malsegregation,37,39 but other data suggest that most abnormalities would be derived from chromatid errors.40,41
In our study, the number of chromosome and chromatid errors was very similar for chromosomes X, 13, 16, 17 and 22 but not for the others. Chromosomes 18 and 21 had more chromatid alterations. Perhaps there is a tendency to precocious chromatid separation in these chromosomes. In unfertilised oocytes, the major mechanism of trisomy formation was chromatid predivision rather than chromosome nondisjunction of whole bivalents.42 Although it was suggested that the high incidence of chromatid predivision was because of the in vitro culture of the oocytes,43 recently the chromatid predivision hypothesis42 has been confirmed in fresh, noninseminated oocytes.38 Balanced predivision has not been considered as a form of aneuploidy,8,38,43 but it should be considered a predisposing factor for aneuploidy.
Our results show more balanced predivision in 1PB than in the corresponding MII (P<0.0001). The human 1PB undergoes rapid degeneration during in vitro incubation. Balanced predivision in 1PBs could be caused by this degenerative process, as suggested.43
We found balanced predivision for all chromosomes analysed (Table 4), although no significant differences were observed. A balanced predivision for chromosome 16 has been reported in 1PBs from unfertilised23,37 and fresh oocytes.38,43 Chromatid predivision of chromosome 1837 and chromosomes 2, 7, 8, 10, 11, 17, 19, 20, 21 and 2238 has also been previously reported. Recent studies in fresh oocytes38 report an increase in balanced predivision with decreasing chromosome size and increasing maternal age.
Aneuploidy rate
The percentage of aneuploid oocytes found in the present study was 47.5%, with 21.25% of hyperhaploidies and 26.25% of estimated nonartefactual hypohaploidies. This is a high frequency of aneuploidy compared with other studies but, in previous publications, a maximum of seven chromosomes were analysed (we analysed nine chromosomes), and the processes to which the oocytes were subjected to in each case were different (they include unfertilised, in vivo and in vitro matured oocytes). On the other hand, by testing the first and second PBs using probes for chromosomes 13, 16, 18, 21 and 22, 45.2% of oocytes were found to be aneuploid, of which 36.1% were of first meiotic origin and 29.3% of second meiotic origin.40 These figures are very similar to ours.
The approximate estimate based on the aneuploidy rate detected for chromosomes 18 and X in unfertilised oocytes was as high as 36.8%,10 and for chromosomes 1, 7, 13, 18, 21 and X it was 44%.44 Summarising data obtained from different studies in which chromosomes were classified by groups, it was found that the mean frequency of aneuploidy obtained in unfertilised and noninseminated oocytes was 22.8%.45 In noninseminated oocytes, the mean frequency of aneuploidy was 37.3%, ranging from 25 to 57.1%.46,47 Other authors found that only 4.7% of oocytes had additional material.37
Recently, testing the 1PBs for chromosomes 13, 16, 18, 21 and 22 in fresh oocytes from women 37–41 years old, 38.2% were found to be aneuploid.26 The use of in vivo matured oocytes from unstimulated cycles, fixing 1PB and the corresponding MII immediately after follicular puncture, would give a better knowledge of oocyte aneuploidy rates.
We found aneuploidy in 39 1PBs from women ranging in age from 28 to 45 years (mean 34.53). In all, 17 aneuploid cells were from women ≥35 years old (mean 37.47 years) and 22 from women <35 years old (mean 31.59). An ANOVA and a Student's t-test showed no significant differences between age and the type of abnormalities (chromosome or chromatid alterations) or the frequency of aneuploidy observed. The sample size of our study was probably too small to demonstrate age-related aneuploidy.
All the patients included in this study underwent ovarian stimulation and most of the oocytes (87.6%) were cultured in vitro during a minimum of 24 h after follicular puncture for in vitro maturation.
It has been suggested48 that ovarian stimulation, which increases the number of recovered oocytes, could induce the maturation of chromosomally abnormal gametes, which would become atrectic without stimulation. The same author reported that aberrations in the number of chromosomes in an oocyte were not only caused by nondisjunction but also by anaphase lag. Human oocytes recovered for in vitro fertilisation seemed to have an incidence of numerical chromosome abnormalities of about 40–50%.
Using probes for chromosomes 16, 18, 21 and X, nondisjunction and chromatid predivision was identified in in vitro matured oocytes from unstimulated ovaries,49 but the authors suggested that hormonal stimulation of the ovary might influence the meiotic nondisjunction rate. However, others50,51 found that superovulation protocols used in IVF might not be responsible for the higher rate of aneuploidy in human oocytes.
The in vitro maturation of the oocytes could artefactually increase the number of aneuploidies observed.49 Other authors43 stated that unbalanced predivision and nondisjunction did not increase significantly with time in culture, but they found an increase in balanced predivision of chromatids also observed previously.8
Some studies52,53 revealed that the spindles of human oocytes were sensitive to prolonged in vitro culture and were morphologically altered. From published results,54 it is conceivable that oocytes from older individuals could be sensitive to even a short culture time, but the morphological anomalies seen in the meiotic spindle suggest other mechanisms, for example, a suboptimal environment for oocyte maturation.
Aneuploidy mechanisms
A multistep model has been proposed as a cause of aneuploidy.55 First, an age-independent step would be needed in the prenatal stage, in which a bivalent susceptibility would be established. The second ‘hit’ would be age-dependent and would involve the abnormal processing of that bivalent, resulting in a decreased efficiency to segregate susceptible bivalents. The effect of hormonal stimulation on susceptible bivalents could increase the probability of obtaining abnormal oocytes that, spontaneously, would have become atrectic. This theory would explain why young women may show a high rate of aneuploidy when their oocytes are obtained by hormonal induction.
A compromised microcirculation in follicles because of the perimenarcheal and perimenopausal hormonal disturbances could lead to a low oxygen concentration that could account for an increased aneuploidy incidence while decreasing the size of the spindle.56 High frequencies of chromosome abnormalities because of segregation disorders in oocytes from severe hypoxic follicles have also been found.57
It is not only age-dependent factors that play a role in the production of aneuploid oocytes; an individual predisposition originating during early meiotic stages could also be very important. Synaptic errors between homologous chromosomes,58 a reduction in the number of chiasmata because of a low recombination rate59 and the subsequent segregation of these unstable bivalents,60 an altered DNA repair because of defects in sister chromatid cohesion61 and other mechanisms are being found to play an important role. Alterations in meiosis-specific proteins involved in the maintenance of the cohesion of sister chromatids55 and mutations in genes required for chiasma formation and for the structural integrity of meiotic bivalents62 have been related to aneuploidy. A lower concentration of transcripts of spindle attachment checkpoint genes in oocytes of older women has also been described.63
In oogenesis, the cell cycle checkpoint mechanism that controls the alignment of chromosomes at metaphase I is not as strict as in the male.64 While in the male, abnormal meiosis is arrested, producing sterility or giving rise to the production of diploid sperm,21 in the female, meiosis is not arrested, and this may give rise to aneuploidies.65 This may explain the higher rate of aneuploidy in female meiosis.49
Focusing on the influence of age in aneuploidy, it was found that in naturally cycling older women (40–45 years old), the meiotic spindle showed many more anomalies affecting chromosome alignment and microtubule matrix.54 These results agree with the incidence of chromosome aneuploidy in karyotyped oocytes in naturally cycling women50 and with the high rate of aneuploidy in older individuals.23
Presence of extra chromosomes
In 25.7% of the patients, oocytes with extra chromosomes were detected. Years ago, some authors8,43 considered this kind of results as FISH errors but, more recently, by analysing embryonic cells66 and analysing oocytes37 these abnormalities were also detected. To better evaluate this phenomenon, a higher number of patients would have to be included and more than one oocyte would have to be analysed from each of them.
The fact that three of these alterations corresponded to the same patient points to an individual predisposition to errors during the proliferative stage in early embryonic development. Some authors37 detected two oocytes with extra chromosomes in one of the patients analysed; these cells were considered to have originated from trisomic germ cell lines. Since in our patient the affected chromosomes were different, we believe that their origin could be found in segregation errors, either unique, multiple or successive, during the mitotic divisions of the precursor oogonia rather than in the existence of trisomic germ cell lines. The lack of relation between age and this type of abnormality in our study is in accordance with this hypothesis.
The mitotic errors mentioned could affect one or more chromosomes; actually, we detected up to four chromosomes involved in the same cell (Table 7). Chromosome 13, alone or with other chromosomes, was the most frequently involved in these oocytes (50%) followed by chromosome 17 (33.3%). The reason why these chromosomes are more implicated is unknown. More cells must be analysed to confirm these frequencies.
The fact that not all oocytes analysed in some patients were affected suggests the existence of chromatid malsegregation during a mitotic anaphase stage in some oogonia that would produce trisomic/nullisomic and disomic germ cells for some chromosomes. This is confirmed by the fact that four of these patients became pregnant during the IVF cycle in which we performed the analysis and all of them gave birth to healthy children. Even patient S, in whom three out of five oocytes had extra chromosomes, gave birth to twins. Regarding the other patients, two of them had miscarriages (one was a t(13;14) carrier and also miscarried in a previous cycle) and three did not become pregnant (one of them had suffered a miscarriage in a previous IVF cycle).
In conclusion, we can say that in spite of the technical difficulties and the partial information obtained, the analysis of 1PBs with nine chromosome probes to select euploid oocytes is a reliable method and it could help advanced maternal age patients undergoing IVF treatments.
References
Meldrum D : Female reproductive aging – ovarian and uterine factors. Fertil Steril 1993; 59: 1–5.
Krey L, Grifo JA : Poor embryo quality: the answer lies (mostly) in the egg. Fertil Steril 2001; 75: 466–468.
Hassold T, Chiu D : Maternal age-specific rates of numerical chromosome abnormalities with special reference to trisomy. Hum Genet 1985; 70: 11–17.
Plachot M, Veiga A, Montagut J et al: Are clinical and biological parameters correlated with chromosomal disorders in early life: a multicentric study. Hum Reprod 1988; 3: 627–635.
Navot D, Bergh P, Williams M et al: Poor oocyte quality rather than implantation failure as a cause of age-related decline in female fertility. Lancet 1991; 337: 1375–1377.
Angell RR, Xian J, Keith J : Chromosome anomalies in human oocytes in relation to age. Hum Reprod 1993; 8: 1047–1054.
Verlinsky Y, Cieslak J, Rechitsky S et al: Safety and accuracy of preimplantation genetic diagnosis. Abstracts Book 2000; Abstract S35: 114–119. 10th International Conference on Prenatal Diagnosis and Therapy.
Dailey T, Dale B, Cohen J, Munné S : Association between nondisjunction and maternal age in meiosis-II. Am J Hum Genet 1996; 59: 176–184.
Munné S, Alikani M, Tomkin G, Grifo J, Cohen J : Embryo morphology, developmental rates and maternal age are correlated with chromosome abnormalities. Fertil Steril 1995; 64: 382–391.
Dyban A, Freidine M, Severova E, Cieslak J, Ivakhnenko V, Verlinsky Y : Detection of aneuploidy mi human oocytes and corresponding first polar bodies by fluorescent in situ hybridization. J Assist Reprod Genet 1996; 13: 73–78.
Márquez C, Cohen J, Munné S : Chromosome identification in human oocytes and polar bodies by spectral karyotyping. Cytogenet Cell Genet 1998; 81: 254–258.
Petit C, Martel-Petit V, Fleurentin A, Monnier-Barbarino P, Joveaux P, Gerard H : Use of PRINS for preconception screening of polar bodies for common aneuploidies. Prenat Diagn 2000; 20: 1067–1071.
Plachot M, Mandelbaum J, Junca A : Qualite de l'oocyte et de l'embryon et resultat de la FIV. Contraception Fert Sex 1990; 18: 636–638.
Ménezo Y, Hazaout A, Dumont M, Herbaut N, Nicollet B : Coculture of embryos on Vero cells and transfer of blastocysts in humans. Hum Reprod 1992; 7: 101–106.
Gianaroli L, Magli MC, Ferraretti AP : The in vivo and in vitro efficiency and efficacy of PGD for aneuploidy. Mol Cell Endocrinol 2001; 183: S13–S18.
Harper JC, Coonen E, Handyside AH, Winston RM, Hopman AH, Delhanty JD : Mosaicism of autosomes and sex chromosomes in morphologically normal, monospermic preimplantation human embryos. Prenat Diagn 1995; 15: 41–49.
Sandalinas M, Sadowy S, Alikani M, Calderon G, Cohen J, Munné S : Developmental ability of chromosomically abnormal human embryos to develop to the blastocyst stage. Hum Reprod 2001; 16: 1954–1958.
Munné S, Magli C, Bahçe M et al: Preimplantation diagnosis of the aneuploidies most commonly found in spontaneous abortions and live births: XY, 13, 14, 15, 16, 18, 21, 22. Prenat Diagn 1998; 18: 1459–1466.
Munné S, Magli C, Cohen J et al: Positive outcome after preimplantation diagnosis of aneuploidy in human embryos. Hum Reprod 1999; 14: 2191–2199.
Gianaroli L, Magli MC, Ferraretti A, Munné S : Preimplantation diagnosis for aneuploidies in patients undergoing in vitro fertilization with a poor prognosis: identification of the categories for which it should be proposed. Fertil Steril 1999; 72: 837–844.
Egozcue J, Santaló J, Giménez C et al: Preimplantation genetic screening and human implantation. J Reprod Immunol 2002; 55: 65–72.
Nicolaidis P, Petersen M : Origin and mechanisms of non-disjunction in human autosomal trisomies. Hum Reprod 1998; 13: 311–319.
Angell RR, Xian J, Keith J, Ledger W, Baird DT : First meiotic division abnormalities in human oocytes: mechanism of trisomy formation. Cytogenet Cell Genet 1994; 65: 194–202.
Durban M, Benet J, Boada M et al: PGD in female carriers of balanced Robertsonian and reciprocal translocations by first polar body analysis. Hum Reprod Update 2001; 7: 591–602.
Munné S, Sapulveda S, Balmaceda J et al: Selection of the most common chromosome abnormalities in oocytes prior to ICSI. Prenat Diagn 2000; 20: 582–586.
Petit C, Bergère M, Lombroso R, Monnier-Barbarino P, Selva J, Gérard H : First pregnancies after polar body biopsy laser-assisted and FISH analysis. Hum Reprod 2002, 17 (Abstracts book 1): 59–60.
Kuliev A, Cieslak J, Ilkevitch Y, Verlinsky Y : Nuclear abnormalities in a series of 6733 human oocytes. Reprod BioMed Online 2002; 4 (Suppl 2): 11. Abstracts Fourth International Symposium on Preimplantation Genetics.
Verlinsky Y, Cieslak J, Ivakhnenko V et al: Prevention of age-related aneuploidies by polar body testing of oocytes. J Assist Reprod Genet 1999; 16: 165–169.
Bahçe M, Cohen J, Munné S : Preimplantation genetic diagnosis of aneuploidy: were we looking at the wrong chromosomes? J Assist Reprod Genet 1999; 16: 176–181.
Barri P, Coroleu B, Martínez F, Veiga A : Stimulation protocols for poor responders and aged women. Mol Cell Endocrinol 2000; 166: 15–20.
Montag M, Van der Ven K, Van der Ven H, Delacretaz G, Rink K : Laser assisted opening of the zona pellucida facilitates polar body biopsy. Hum Reprod 1997; 12: 10. Abstracts of the 13th Annual Meeting of ESHRE.
Durban M, Benet J, Sarquella J, Egozcue J, Navarro J : Chromosome studies in first polar bodies from hamster and human oocytes. Hum Reprod 1998; 13: 583–587.
Tarkowski A : An air drying method for chromosome preparations from mouse eggs. Cytogenetics 1966; 5: 394–400.
Drury K, Kovalinskaia L, Clark P, Zhaoheng D, Williams R : Novel 5 minute hybridization procedure for FISH preimplantation diagnosis. Fertil Steril 1997; n. (Suppl 1). Abstracts 53rd Annual Meeting, p S50.
Bahce M, Escudero T, Sandalinas M, Morrison L, Legator M, Munné S : Improvements of preimplantation diagnosis of aneuploidy by using microwave hybridization, cell recycling and monocolour labelling of probes. Mol Hum Reprod 2000; 6: 849–854.
Hassold T, Jacobs P : Trisomy in man. Annu Rev Genet 1984; 18: 69–97.
Mahmood R, Brierley C, Faed M, Mills J, Delhanty J : Mechanisms of maternal aneuploidy: FISH analysis of oocytes and polar bodies in patients undergoing assisted conception. Hum Genet 2000; 106: 620–626.
Sandalinas M, Márquez C, Munné S : Spectral karyotyping of fresh, non-inseminated oocytes. Mol Hum Reprod 2002; 8: 580–585.
Verlinsky Y, Cieslak J, Freidine M et al: Pregnancies following pre-conception diagnosis of common aneuploidies by fluorescent in-situ hybridization. Hum Reprod 1995; 10: 1923–1927.
Verlinsky Y, Cieslak J, Ivakhenko V et al: Chromosomal abnormalities in the first and second polar body. Mol Cell Endocrinol 2001; 183 (Suppl 1): S47–S49.
Angell RR : First meiotic division nondisjunction in human oocytes. Am J Hum Genet 1997; 61: 23–32.
Angell RR : Predivision in human oocytes at meiosis I: a mechanism for trisomy formation in man. Hum Gene 1991; 86: 383–387.
Munné S, Dailey T, Sultan K, Grigo J, Cohen J : The use of first polar bodies for preimplantation diagnosis of aneuploidy. Hum Reprod 1995; 10: 1014–1020.
Martini E, Flaherty SP, Swann NJ, Matthews CD, Ramaekers F, Geraedts JP : FISH analysis of six chromosomes in unfertilized human oocytes after polar body removal. J Assist Reprod Genet 2000; 17: 276–283.
Pellestor F : Frequency and distribution of aneuploidy in human female gametes. Hum Genet 1991; 86: 283–288.
Wramsby H, Liedholm P : A gradual fixation method for chromosomal preparations of human oocytes. Fertil Steril 1984; 41: 736–738.
Martin R, Mahadevan M, Taylor P et al: Chromosomal analysis of unfertilized human oocytes. Reprod Fertil 1986; 78: 673–678.
Wramsby H, Fredga K, Liedholm P : Chromosome analysis of human oocytes recovered from preovulatory follicles in stimulated cycles. N Engl J Med 1987; 316: 121–124.
Volarcik K, Sheean L, Goldfarb J, Woods L, Abdul-Karim F, Hunt P : The meiotic competence of in-vitro matured human oocytes is influenced by donor age: evidence that folliculogenesis is compromised in the reproductively aged ovary. Hum Reprod 1998; 13: 154–160.
Gras L, McBain J, Trounson A, Kola I : The incidence of chromosomal aneuploidy in stimulated and unstimulated (natural) uninseminated human oocytes. Hum Reprod 1992; 7: 1396–1401.
Zenzes M, Wang P, Casper R : Evidence for maternal predisposition to chromosome aneuploidy in multiple oocytes of some in vitro fertilisation patients. Fertil Steril 1992; 57: 143–149.
Eichenlaub-Ritter U, Stahl A, Luciani J : The microtubular cytoskeleton and chromosomes of unfertilised human oocytes aged in vitro. Hum Genet 1988; 80: 259–264.
Pickering S, Johnson M, Braude P, Houliston E : Cytoskeletal organization in fresh, aged and spontaneously activated human oocytes. Hum Reprod 1988; 3: 978–989.
Battaglia DE, Goodwin P, Klein NA, Soules MR : Influence of maternal age on meiotic spindle assembly in oocytes from naturally cycling women. Hum Reprod 1996; 11: 2217–2222.
Lamb N, Freeman S, Savage-Austin A et al: Susceptible chiasmate configurations of chromosome 21 predispose to nondisjunction in both maternal meiosis-I and meiosis-II. Nat Genet 1996; 14: 400–405.
Gaulden M : A model that explains the varying frequency of aneuploid children with maternal age (J-shaped curve) as well as aneuploidy of paternal origin. Prog Clin Biol Res 1989; 318: 253–257.
Van Blerkom J, Antczak M, Schrader R : The developmental potential of the human oocyte is related to the dissolved oxygen content of follicular fluid: association with vascular endothelial growth factor levels and perifollicular blood flow characteristics. Hum Reprod 1997; 12: 1047–1055.
Vidal F, Navarro J, Templado C, Egozcue J : Synaptonemal complex studies in the male. Hum Reprod 1987; 2: 577–581.
Hassold T, Hunt P : To err (meiotically) is human: the genesis of human aneuploidy. Nature 2001; 2: 280–290.
Wolstenholme J, Angell RR : Maternal age and trisomy–a unifying mechanism of formation. Chromosoma 2000; 109: 435–438.
Nasmyth K : Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu Rev Genet 2001; 35: 673–745.
Yuan L, Liu J, Hoja M, Wilbertz J, Nordqvist K, Höög C : Female germ cell aneuploidy and embryo death in mice lacking the meiosis-specific protein SCP3. Science 2002; 296: 1115–1118.
Steuerwald N, Cohen J, Herrara R, Sandalinas M, Brenner C : Association between spindle assembly checkpoints expression and maternal age in human oocytes. Mol Hum Reprod 2001; 7: 49–55.
Hunt P, LeMaire R, Embury P, Sheean L, Mroz K : Analysis of chromosome behaviour in intact mammalian oocytes: monitoring the segregation of a univalent chromosome during female meiosis. Hum Mol Genet 1995; 4: 2007–2012.
Eichenlaub-Ritter U, Cucurkam S, Betzendahl I, Yin H : Studies on the aneugenic properties of trichlorfon, a pesticide, vermicide and drug used in the treatment of Alzheimer patients. Hum Reprod 1999; 14: 240–241.
Conn CM, Cozzi J, Harper JC, Winston RM, Delhanty JD : Preimplantation genetic diagnosis for couples at high risk of Down syndrome pregnancy owing to parental translocation or mosaicism. J Med Genet 1999; 36: 45–50.
Acknowledgements
This work was supported by Ministerio de Sanidad (FIS 98/0031-01) (FIS 2002/Pi020168), CIRIT (1997 SGR-00030) and Fundació Catalana Síndrome de Down/Marató de TV3 (1994–98). It has also been supported by Departament d'Universitats, Recerca i Societat de la Informació de la Generalitat de Catalunya (Aïda Pujol Masana has a Beca Predoctoral de Formació de Personal Investigador; FI/FIAP) and by the Càtedra de Investigació en Obstetrícia i Ginecologia de l'Institut Universitari Dexeus.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Pujol, A., Boiso, I., Benet, J. et al. Analysis of nine chromosome probes in first polar bodies and metaphase II oocytes for the detection of aneuploidies. Eur J Hum Genet 11, 325–336 (2003). https://doi.org/10.1038/sj.ejhg.5200965
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5200965