Abstract
The incidence of Y/autosome translocations is low. Whereas involvement of non-acrocentric chromosomes often leads to infertility, cases related with acrocentric chromosomes are usually familial with no or minimal effect on fertility. A de novo (Yp/13p) translocation was found in a 32-year-old male referred for severe oligozoospermia. Conventional cytogenetic procedures (GTG, CBG and NOR banding) and molecular cytogenetic techniques (Fluorescence In Situ Hybridization, FISH) were performed on high-resolution chromosomes obtained after peripheral blood lymphocyte culture as also on interphase nuclei of spermatogenic cells from semen samples. Screening of AZF microdeletions in the Yq11.2 region known to be involved with spermatogenesis defects was also performed. GTG banding showed a (Yp/13p) translocation in all scored metaphases. CBG and NOR staining of the derivative chromosome revealed the maintenance of Yq heterochromatin and of the 13p NOR region. FISH with centromeric Y and 13/21 probes, SRY specific probe and X/Y (p and q arms) sub-telomeric probes gave the expected number/location of fluorescent signals. Hybridisation with a pan-telomeric repeat (TTAGGG) probe showed an absence of the telomeric sequences at the fusion point of the rearranged chromosome. FISH analysis with probes to chromosomes X, Y, 13 and 18 showed an abnormal segregation of the translocated chromosome during meiosis I, which explains that only 13.6% of the secondary spermatocytes were normal. Most of these became arrested, as after meiosis II the large majority of the round spermatids were normal (70%), as were in consequence most of the sperm (85.1%). Multiplex-PCR confirmed the intactness of the SRY region and showed absence of AZF microdeletions. We report a novel de novo (Yp;13p) translocation characterised by loss of the 13p and Yp telomeres. Meiotic studies using FISH demonstrated meiosis I chromosome unpairing and mal segregation that justifies the severe oligozoospermia. Although most sperm have a normal chromosomal constitution, preimplantation genetic diagnosis should be considered an option for this patient.
Similar content being viewed by others
Introduction
The frequency of Y/autosome translocations in the general population is approximately 1 in 2000.1,2,3 Like any other chromosome, the Y chromosome can be translocated onto an autosome, an X chromosome or another Y chromosome, either in a balanced or unbalanced way. Translocations between the Y and a non-acrocentric chromosome are rare and may involve any part of the Y chromosome,4 often leading to an abnormal phenotype and infertility.4,5,6,7,8,9 However, in the most common form of Y/autosome translocation, the heterochromatic portion of Yq is translocated onto the short arm of an acrocentric chromosome.10 These translocations have been observed in phenotypically normal individuals and reported in multiple families, indicating that fertility is usually not affected.4,10,11,12
In the last 5 years, there were two new reports on (Y;13) translocations, both involving breakpoints on the short arms of chromosomes Y and 13 and with evident loss of chromosomal material.13,14 In this study we describe a novel de novo (Yp;13p) translocation found in a 32-year-old man referred for severe oligozoospermia.
Methods
Informed consent was obtained from the patient and his relatives before inclusion in the study.
Clinical findings
A phenotypically normal 32-year-old male, 170 cm in height, was referred to infertility treatment due to severe oligozoospermia. In his past history the patient experienced epidemic parotiditis at age 10, apparently associated with bilateral orchitis. Clinical examination revealed atrophy of the left testicle and a slightly hypotrophic right testicle, with normal epididymis and vas deferens. Hormone serum levels were 10 mU/ml FSH, 2.1 mU/ml LH and 3.2 ng/ml testosterone. The spermiogram, after centrifugation, showed only a few immobile spermatozoa.
Karyotyping and fluorescence in situ hybridisation
High-resolution chromosomal GTG banding, as also CBG and NOR banding to characterise the translocation breakpoints, were performed on cultured, PHA-stimulated peripheral blood lymphocytes, according to standard cytogenetic procedures.15 Fluorescence In Situ Hybridisation (FISH) followed manufacturer's instructions, using centromeric probes for chromosomes Y (DYZ3, Yp11.1-q11.1; Vysis) and 13/21 (D13Z1/D21Z1, 13p11.1-q11.1 and 21p11.1-q11.1; Cytocell), a SRY-specific probe (SRY, Yp11.3; Vysis), subtelomere-specific probes for both arms of the sex chromosomes (DXS28, Xp/Yp telomeres; DXS7059, Xq/Yq telomeres; Cytocell) and a probe for the telomeric repeats of all chromosomes (STAR FISH, Perbio, Cambio). A 13p telomeric probe was not used due to its unavailability. Normal male lymphocyte preparations were used as controls for the FISH assays.
Meiotic studies
Fresh semen samples were centrifuged at 1500 r.p.m. (900 g), 10 min, at room temperature, to wash out the seminal fluid. The pellet was washed three times in PBS by centrifugation, fixed (5 min) with methanol-glacial acetic acid (3 : 1), centrifuged, and resuspended in three drops of the fixative. Ten μl of this suspension were transferred to poly-L-lysine coated slides, air-dried, incubated (5 min) in 2×SSC to remove the remainders of the fixative, dehydrated in ethanol (70, 96 and 100%, 5 min in each), and air-dried before adding the probes. For analysis of mature sperm, some slides were incubated (10 min) with 1 M DTT in 1 M Tris buffer, pH 9.5, to make the condensed chromatin accessible to probes, washed in 2×SSC (5 min) and PBS (5 min), and finally dehydrated as above.16 For FISH, a locus specific probe for chromosome 13 (13q14, Vysis) and α-satellite probes for chromosomes Y (DXZ3, Yp11.1–q11.1, Vysis), X (DXZ1, Xp11.1–q11.1, Vysis) and 18 (D18Z1, 18p11.1–q11.1, Vysis) were used.17 Normal male lymphocyte preparations were used as controls for the efficiency of the FISH procedure. Classification of germ cells was made using inverted Hoffman microscopy and after fluorescent in situ hybridization was mainly based on the size and density of the cell nucleus.18
PCR amplification for SRY and AZF regions
Peripheral blood samples (8–10 ml) were pelleted and stored at −20°C until DNA extraction. High molecular weight DNA was isolated using a salting-out method. The patient was screened for Yq11.2 chromosome microdeletions in the AZF regions by multiplex-PCR using six STS markers (AZFa: sY84; AZFb: sY134, sY142; AZFc: sY152, sY157, sY254). SRY (Yp) was used as an internal positive control. Negative controls included normal male and female DNA and PCR reaction with no DNA added. As no deletion was found, new additional markers were screened in each region (AZFa: sY86; GY6, DFFRY; AZFb: sY114, sY127, sY135; AZFc: sY153, sY255, DAZ1, CDY1). A STS was considered absent or negative after three amplification failures in independent PCR reactions.19 The present protocol was certified by EAA and EMQN (EQAS Y Chromosome-Ref. Nr.: 01; 9 May 2001).
Results
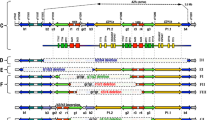
In all 30 observed metaphases, GTG banding showed that the entire Y chromosome was translocated onto one of the chromosomes 13, with breakpoints on their short arms (Figure 1A,D). The patient's father and only brother had a normal karyotype (46,XY), demonstrating that this was a de novo (Yp;13p) translocation. A normal staining pattern of all heterochromatic segments, including the distal Yqh, was shown by CBG banding (Figure 1B,D). NOR banding showed that the NOR region was conserved on the translocation chromosome, suggesting a breakpoint on 13p12 (Figure 1C,D).
FISH analysis with centromeric probes for chromosomes Y (Figure 2A) and 13/21 (Figure 2B) confirmed that the derivative (Y;13) chromosome was dicentric. The intactness of the SRY locus on the Yp arm showed that the Yp11.2 region was present (Figure 2C), as were also the telomeric specific regions of both arms of the sex chromosomes (Figure 2D), since hybridisation with subtelomeric probes for Xp/Yp and Xq/Yq gave positive signals at the free end of the Y chromosome and also at the breakpoint fusion of the derivative chromosome. Hybridization with a pantelomeric repeat probe showed that the Y and 13 chromosomes had lost their p-telomeres, thus defining the breakpoint on the Yp11.32 band (Figure 2E). In the control slides of the FISH procedure, 95–98% of the cells gave correct hybridisation signals for each of the probes used. The karyotype was then interpreted as:
(A) Y centromeric probe (red signal) hybridising on the rearranged (Y;13). (B) 13/21 centromeric probe (yellow signals) hybridising on both chromosomes 13 and 21. (C) SRY specific probe (red signal) binding the SRY gene on the derivative (Y;13). (D) Subtelomere specific probes binding to both arms (p- yellow signal; q- red signal) of the sex chromosomes. (E) Pantelomeric repeat probe binding to the free ends of all chromosomes, with no staining at the fusion point of the derivative chromosome. Arrows indicate the derivative chromosome.

Molecular studies confirmed the intactness of the SRY on Yp and demonstrated absence of microdeletions in the Yq11.2-AZF regions (Figure 3).
FISH analysis of immature germ cells and mature sperm (Tables 1, 2; Figure 4) showed a predominance of chromosome unpairing (73.5%) in pre-meiotic diploid germ cells and an abnormal chromosome segregation during meiosis I due to the presence of the translocation (56.8%). On the contrary, most of the round spermatids (70%) and sperm (85.1%) were normal for the referred chromosomes. In control slides, 93% of the cells gave correct hybridisation signals for the probes used.
Hybridisation with probes for chromosomes 13 (yellow), 18 (orange), X (Magenta) and Y (red). (A) Primary spermatocyte with normal pairing. (B) Primary spermatocyte with unpaired X and 18 chromosomes. (C) Normal secondary spermatocyte. (D) Round spermatids with aneuploidies resulting from seggregation errors. (E) Spermatozoa showing normal chromosome complement. (F) Spermatozoon with hiperploidy.
Discussion
To our knowledge, there are only two cases reported in the literature describing a (Yp;13p) translocation. One regards a 13-year-old boy with moderate psychomotor retardation, mild facial asymmetry, dental malocclusion and short fifth fingers and toes. After GTG banding, a de novo (Y;13) translocation was revealed in 90% of the cells, with an apparently intact Y chromosome, including the SRY and PAR-regions, and a chromosome 13 that had lost part of the short arm. Hybridisation with telomeric probes also showed that these sequences were present at each free telomeric extremity and at the fusion point of the (Y;13).13 The second case is related to a 10-year-old boy with poor growth since birth, but with normal psychomotor development. GTG banding showed an abnormal male karyotype, 45,X,dic(Y;13)(p1?;p1?), and FISH studies with centromeric probes for chromosomes Y and 13/21 confirmed a dicentric derivative chromosome. PCR and dosage Southern analysis revealed the loss of the 13p NOR and satellite-regions and of the entire PAR-region on Yp.14
We report here a new type of translocation involving the short arms of Y and 13 chromosomes. Our subject had normal development with an otherwise normal phenotype besides presenting a severe oligozoospermia. The cytogenetic analysis of GTG banded chromosomes revealed a de novo (Y;13) translocation in all metaphase cells. The translocation breakpoints were clearly defined after NOR and FISH studies, which showed that the translocated chromosome 13 had intact NOR regions and had lost only a very small part of its short arm (satellite heterochromatin and telomere), whereas the Y chromosome apparently lost only the p-telomere. The pseudoautosomal region (PAR) of the sex-chromosomes is localised at both the XpYp and XqYq ends, and in this particular case the presence of specific sub-telomeric sequences suggests that the PAR1-region was not affected.
During meiosis, the sex chromosomes form a XY body or ‘sex vesicle’ and are asynchronously condensed. In the case of an X or Y/autosome translocation, a translocated chromosome will be included in the compartment of the XY body and inactivation may extend to the autosomal segment yielding degeneration of most spermatocytes after the pachytene stage.21,22 The PAR1 (at XpYp) represents the region for sex-chromosome recombination and must be intact in order to promote meiotic pairing and ensure sperm production.23,24,25 However, the fact that the PAR 1 region is intact does not necessarily imply that the meiotic pairing is normal, as telomere TTAGGG repeats, rather than subtelomeric sequences, play an important role in meiotic pairing. They are responsible for the movement of telomeres at the surface of the nuclear envelope and the initiation of chromosome pairing at the zygotene stage.26 In the present case, as these telomeric repeats, which normally flank the X and Y PAR-regions, are lacking, the initiation of X and Y pairing may be delayed or disturbed. The presence of a whole chromosome 13 adjacent to the Y chromosome may also restrict the accessibility of the Y-PAR region and then alter the formation of the XY body at pachytene. In addition, pairing of both chromosomes 13, initiated from telomeres, may also interfere with the formation of the XY body.21,22
In the present study, an abnormal segregation of the (Y;13) was found during meiosis I, as only 13.6% of the secondary spermatocytes showed a normal chromosome complement. In this situation, most of the abnormal secondary spermatocytes might become arrested and degenerated,18 which explains why the large majority of the round spermatids were normal (70%), as were in consequence most of the sperm (85.1%). Results also evidenced unpaired chromosome centromeric signals in most (73.5%) of the diploid germ cells, thus suggesting that the majority of the cells might not overcome meiosis I, blocking and degenerating at the primary spermatocyte stage. However, besides the sex chromosomes and the 13 chromosome, unpairing was also extensive to chromosome 18, which excludes specificity for the translocated chromosome. If in the present case there was a predominance of round spermatogonia and of preleptotene/leptotene spermatocyte stages this could be a normal finding.26 Notwithstanding, the study of the products of meiosis I clearly reveal that an abnormal segregation (and thus pairing) of the (Y;13) chromosome occurs, as most of the secondary spermatocytes were abnormal due to the translocated chromosome (56.8%).
In conclusion, the presence of an intact PAR-region, absence of Y chromosome microdeletions in the AZF-regions, absence of the telomere repeat sequences at the fusion point and the FISH meiotic studies indicate that the severe oligozoospermia may be due to the translocation, through a disturbance in meiotic pairing and segregation, rather than to the past orchitis. The observation of a high percentage of spermatozoa with an apparently normal chromosomal content (85.1%), almost as high as in the general population,18 is of very good prognosis for this patient, since the selection of these cells for ICSI does not represent an increased risk for aneuploidies. However, selection of female embryos by PGD should be considered at genetic counselling.
References
Nielsen J, Rasmussen K . Y/autosomal translocations Clin Genet 1976 9: 609–617
Powell C . Sex chromosomes and sex chromosomes abnormalities in Gersen SL, Keagle MB (eds) The Principles of Clinical Cytogenetics New Jersey, Totowa: Humana Press 1984 pp 229–258
Gardner RJM, Sutherland GR . Chromosome Abnormalities and Genetic Counseling. Sex chromosome translocations New York, Oxford: Oxford University Press 1996 pp 95–114
Smith A, Fraser IS, Elliot G . An infertile man with balanced Y;19 translocation. Review of Y;autosome translocations Ann Genet 1979 22: 189–194
Delobel B, Djlelati R, Gabriel-Robez O, Croquete M-F . Y-autosome translocation and infertility: usefulness of molecular, cytogenetic and meiotic studies Hum Genet 1998 102: 98–102
Goodfellow P, Darling S, Wolfe J . The human Y chromosome J Med Genet 1985 22: 324–344
Vogt PH, Edelmann A, Kirsch S et al. Human Y chromosome azoospermia factors (AZF) mapped to different subregions in Yq11 Hum Mol Genet 1996 5: 933–943
Vogt PH . Y chromosome and single gene deffects that cause male infertility in: Jansen R, Mortimer D (eds) Towards reproductive certainty: fertility and genetics beyond 1999 Carnforth, UK: Parthenon 1999 pp 298–312
Viguié F, Romani F, Dadoune JR . Male infertility in a case of (Y;6) balanced reciprocal translocation Hum Genet 1982 62: 225–227
Hsu LYF . Phenotype/karyotype correlations of Y chromosome aneuploidy with emphasis on structural aberrations in postnatally diagnosed cases Am J Med Genet 1994 53: 108–140
Cohen MM, Frederick RW, Balkin NE, Simpson SJ . The identification of Y chromosome translocation following distamycin A treatment Clin Genet 1981 19: 335–342
Alitalo T, Tiihonen J, Hakola P, la Chapelle A . Molecular characterization of a Y;15 translocation seggregating in a family Hum Genet 1988 79: 29–35
Boutouil M, Fetni R, Qu J, Dallaire L, Richer C-L, Lemieux L . Fragile site and interstitial telomere repeat sequences at the fusion point of a de novo (Y;13) translocation Hum Genet 1996 98: 323–327
Shanske A, Ellison J, Vuguin P, Dowling P, Wasserman E, Heinrich J, Saenger P . Deletion of the pseudoautosomal region in a male with a unique Y;13 translocation and short stature Am J Med Genet 1999 82: 34–39
Verma RS, Babu A . Human Chromosomes – Principles and Techniques McGraw-Hill 1995 pp 1–183
Coonen E, Pieters MHEC, Dumoulin JCM, Meyer H, Evers JLH, Ramaekers FCS, Geraedts JPM . Non-isotopic in situ hybridization as a method for nondisjunction studies in human spermatozoa Mol Reprod Develop 1991 28: 18–22
Coonen E, Dumoulin JCM, Meyer H, Ramaekers FCS, Hopman AHN . Optimal preparation of Preimplantation embryo interphase nuclei for analysis by fluorescence in situ hybridization Hum Reprod 1994 3: 533–537
Sousa M, Cremades N, Alves C, Silva J, Barros A . Developmental potencial of human spermatogenic cells co-cultured with Sertoli cells Hum Reprod 2002 17: 161–172
Barros A, Carvalho F, Fernandes S, Torgal A, Silva J, Sousa M . Population screening of Y-chromosome microdeletions in Portugal: a study involving 428 infertile male patients Hum Reprod 2001 16: 189
Mitelman F . ISCN 1995. An International System for Human Cytogenetic Nomenclature Basel: S Karger 1995
Delobel B, Djelati R, Gabriel-Robez O et al. Y-autosome translocation and infertility: usefulness of molecular, cytogenetic and meiotic studies Hum Genet 1998 102: 98–102
Guichaoua MR, Quack B, Speed RM, Noel B, Chandley AC, Luciani JM . Infertility in human males with autosomal translocations: meiotic study of a 14;22 robertsonean translocation Hum Genet 1990 86: 162–166
Mohandas TK, Speed RM, Passage MB, Yen PH, Chandley AC, Shapiro LJ . Role of the pseudoautosomal region in sex-chromosome pairing during male meiosis: meiotic studies in a man with a deletion on distal Xp Am J Hum Genet 1992 51: 526–533
Rappold G . The pseudoautosomal regions of the human sex chromosomes Hum Genet 1993 92: 315–324
Gabriel-Robez O, Rumpler Y . The meiotic pairing behaviour in human spermatocyte carriers of chromosome abnormalities and their repercussions on reproductive fitness. II. Robertsonean and reciprocal translocations. A European study Ann Genet 1990 39: 17–25
Scherthan H, Weich S, Schwegler H, Heyting C, Harle M, Cremer T . Centromere and telomere movements during early meiotic prophase of mouse and man are associated with the onset of chromosome pairing J Cell Biol 1996 134: 1109–1125
Acknowledgements
We are grateful to Dr Rafael Bernabeu, Institute Bernabeu, Alicante, Spain, for giving us the opportunity to study this patient. We also thank our laboratory technicians, Mrs Fernanda Lemos and Mrs Filomena Reis for their precious help with the cytogenetic procedures. This study was partially supported by FCT (Sapiens 36363/99, 35231/99; UMIB).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Alves, C., Carvalho, F., Cremades, N. et al. Unique (Y;13) translocation in a male with oligozoospermia: cytogenetic and molecular studies. Eur J Hum Genet 10, 467–474 (2002). https://doi.org/10.1038/sj.ejhg.5200835
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5200835
Keywords
This article is cited by
-
An infertile azoospermic male with 45,X karyotype and a unique complex (Y;14); (Y;22) translocation: cytogenetic and molecular characterization
Journal of Assisted Reproduction and Genetics (2018)
-
Meiotic outcome in two carriers of Y autosome reciprocal translocations: selective elimination of certain segregants
Molecular Cytogenetics (2017)
-
An infertile 45,X male with a SRY-bearing chromosome 13: a clinical case report and literature review
Journal of Assisted Reproduction and Genetics (2015)
-
Mosaicism for an unbalanced Y;21 translocation in an infertile man: a case report
Journal of Assisted Reproduction and Genetics (2013)