Abstract
We report the identification of a novel locus responsible for an autosomal recessive form of hearing loss (DFNB) segregating in a Palestinian consanguineous family from Jordan. The affected individuals suffer from profound prelingual sensorineural hearing impairment. A genetic linkage with polymorphic markers surrounding D9S1776 was detected, thereby identifying a novel deafness locus, DFNB31. This locus could be assigned to a 9q32-34 region of 15 cM between markers D9S289 and D9S1881. The whirler (wi) mouse mutant, characterised by deafness and circling behaviour, maps to the corresponding region on the murine chromosome 4, thus suggesting that DFNB31 and whirler may result from orthologous gene defects.
Similar content being viewed by others
Introduction
Hearing impairment is the most frequent sensorineural defect in man. Approximately one in 1000 children is affected with prelingual deafness. In the majority of cases, deafness is isolated (or nonsyndromic), ie, hearing impairment is the sole symptom. Isolated deafness forms transmitted in the recessive mode are the most frequent (85%) and the most severe. Twenty-six different DFNB loci have been described to date (http://www.uia.ac.be/dnalab/hhh/ and Petit et al1 for review). Eleven of the corresponding genes have been identified (reviewed in Petit et al1 and Verpy et al2). Using the approach based on homozygosity mapping in large affected consanguineous families, we have already identified five DFNB loci in the Lebanese population.3,4,5,6,7 Here, we report on the identification of a novel DFNB locus in a Palestinian family living in Jordan.
Methods
Informed consent was obtained from adult subjects and from parents of under-aged patients.
Auditory tests
Affected as well as unaffected members of the family underwent a general careful clinical and otoscopic examination. Pure tone audiometry with air and bone conduction at 250, 500, 1000, 2000, 4000 and 8000 Hz was performed (with a Beltone 2000 clinical audiometer) in every individual over 5 years of age. Air conduction pure-tone average (ACPTA) threshold in the conversational frequencies (0.5, 1 and 2 kHz) was calculated for each ear, and the hearing loss was classified as mild (20 dB⩽ACPTA⩽40 dB), moderate (40 dB
Genotypying
DNA was prepared from 5–10 ml of blood. Fluorescent microsatellite markers (sequences published in Dib et al8) were used.9 The genome-wide screening was performed using 400 microsatellite markers that are spaced 10–12 cM apart (ABI PRISM® Linkage Mapping Set 2, Applied Biosystems). Multiplex PCR were performed with 4 markers. PCR products were analysed following electrophoresis on a 6% polyacrylamide gel.
Linkage analysis
LOD scores were calculated using the MAPMAKER/HOMOZ program10 (version 0.9), a computer package based on an algorithm especially designed for homozygosity mapping. The defect was assumed to be inherited on a recessive mode and fully penetrant. The disease allele frequency was estimated at 10−3; changing the disease allele frequency to 10−2 only slightly modified the lodscore value. The allele frequencies of the polymorphic markers and the meiotic recombination frequencies for males and females were assumed to be equal.
Results and Discussion
Clinical description
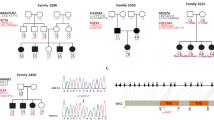
In family JO-CH, four individuals (IV-1, IV-2, IV-3 and IV-8) born from a marriage between two first cousins (III-1 and III-2), were affected by hearing loss (Figure 1). A second consanguineous marriage between first cousins (IV-10 and IV-11) resulted in six children among whom two had been recognised as deaf in the prelingual period. In the six affected children IV-1, IV-2, IV-3, IV-8, V-2 and V-5 aged 32, 29, 27, 13, 11 and 5 years respectively, hearing impairment was bilateral and profound. No air-bone gap was observed in the audiograms, indicating that deafness is sensorineural. No other clinical sign was associated to deafness. Audiometric tests were normal in both parents. It can thus be concluded that a recessive form of isolated hearing loss is segregating in this family.
Segregation of 6 polymorphic markers from the 9q32-34 chromosomal region in family JO-CH. The genetic distance (in cM) between these markers8 is indicated at the bottom left. Horizontal double bars indicate consanguineous unions. Dark symbols represent deaf individuals. Analysis of the haplotypes in generation IV defines the DFNB31 minimal interval as the 15 cM region between D9S289 and D9S1881.
Linkage mapping
We undertook a screening of the whole genome using polymorphic markers spaced 10–12 cM apart.8 A linkage was detected only with locus D9S1776 on chromosome 9, thereby defining a novel deafness locus, DFNB31. Five other polymorphic markers from this chromosomal region were tested (see Figure 1). Two-point lodscores (Zmax) at a recombination fraction θ=0.00 were 5.20, 4.98 and 3.56 for D9S1824, D9S1776 and D9S1682, respectively. The calculated multipoint lodscore was 6.14. The homozygous region common to the six affected individuals extends between D9S1824 and D9S1682. The closest flanking markers limiting the interval are D9S289 and D9S1881, located at q32 and q34, respectively; these markers are spaced about 15 cM.8
Several DFNB forms have been shown to be allelic to dominant forms of isolated deafness (DFNA) and/or to syndromic forms of deafness (see Petit et al1 and Petit11 for reviews). So far, no DFNA form or syndromic hearing loss has been assigned to the DFNB31 chromosomal region.
Among the numerous deaf mouse mutants which have been reported, ten have already been proven to involve genes orthologous to those implicated in human isolated hearing loss (see1,12,13 for reviews). The region syntenic to the DFNB31 interval is located on the murine chromosome 4, 31 cM from the centromere. It contains the locus of the recessive mutant whirler (wi),14,15 which is characterised by profound deafness and vestibular dysfunction. Therefore, it is reasonable to propose that DFNB31 and whirler may represent defects of the same gene.
References
Petit C, Levilliers J, Hardelin J-P . Molecular genetics of hearing loss Annu Rev Genet 2001 35: 589–646
Verpy E, Masmoudi S, Zwaenepoel I et al. Mutations in a new gene encoding a protein of the hair bundle cause non-syndromic deafness at the DFNB16 locus Nature Genet 2001 29: 345–349
Chaïb H, Place C, Salem N et al. A gene responsible for a sensorineural nonsyndromic recessive deafness maps to chromosome 2p22-23 Hum Mol Genet 1996 5: 155–158
Chaïb H, Place C, Salem N et al. Mapping of DFNB12, a gene for a non-syndromal autosomal recessive deafness, to chromosome 10q21-22 Hum Mol Genet 1996 5: 1061–1064
Mustapha M, Chardenoux S, Nieder A et al. A sensorineural progressive autosomal recessive form of isolated deafness, DFNB13, maps to chromosome 7q34-q36 Eur J Hum Genet 1998 6: 245–250
Mustapha M, Salem N, Weil D, El-Zir E, Loiselet J, Petit C . Identification of a locus on chromosome 7q31, DFNB14, responsible for prelingual sensorineural non-syndromic deafness Eur J Hum Genet 1998 6: 548–551
Mustapha M, Weil D, Chardenoux S et al. An a-tectorin gene defect causes a newly identified autosomal recessive form of sensorineural pre-lingual non-syndromic deafness, DFNB21 Hum Mol Genet 1999 8: 409–412
Dib C, Fauré S, Fizames C et al. A comprehensive genetic map of the human genome based on 5,264 microsatellites Nature 1996 380: 152–154
Gyapay G, Ginot F, Nguyen S, Vignal A, Weissenbach J . Genotyping procedures in linkage mapping Methods 1996 9: 91–97
Kruglyak L, Daly MJ, Lander ES . Rapid multipoint linkage analysis of recessive traits in nuclear families, including homozygosity mapping Am J Hum Genet 1995 56: 519–527
Petit C . Usher syndrome: from genetics to pathogenesis Annu Rev Genomics & Hum Genet 2001 2: 271–297
Steel KP . Inherited hearing defects in mice Annu Rev Genet 1995 29: 675–701
Ahituv N, Avraham KB . Auditory and vestibular mouse mutants: Models for human deafness J Basic & Clin Physiol & Pharmacol 2000 11: 181–191
Fleming J, Rogers MJC, Brown SDM, Steel KP . Linkage analysis of the whirler deafness gene on mouse chromosome 4 Genomics 1994 21: 42–48
Rogers MJC, Fleming J, Kiernan BW et al. Genetic mapping of the whirler mutation Mamm Genome 1999 10: 513–519
Acknowledgements
We are grateful to the members of the family who participated in this study. We thank Delphine Torchard-Pagnez and Delphine Bacq for help in genotyping and Jean-Pierre Hardelin and Jacqueline Levilliers for their help in the preparation of the manuscript. This work was supported by grants from Fondation pour la Recherche Médicale (ARS 2000), EC QLG2-CT-1999-00988, and Université Saint-Joseph (Lebanon).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mustapha, M., Chouery, E., Chardenoux, S. et al. DFNB31, a recessive form of sensorineural hearing loss, maps to chromosome 9q32-34. Eur J Hum Genet 10, 210–212 (2002). https://doi.org/10.1038/sj.ejhg.5200780
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5200780
Keywords
This article is cited by
-
Identification of autosomal recessive nonsyndromic hearing impairment genes through the study of consanguineous and non-consanguineous families: past, present, and future
Human Genetics (2022)
-
Genetics, pathogenesis and therapeutic developments for Usher syndrome type 2
Human Genetics (2022)
-
Gene Therapy Restores Hair Cell Stereocilia Morphology in Inner Ears of Deaf Whirler Mice
Molecular Therapy (2016)
-
Analysis of two Arab families reveals additional support for a DFNB2 nonsyndromic phenotype of MYO7A
Molecular Biology Reports (2014)
-
DFNB79: reincarnation of a nonsyndromic deafness locus on chromosome 9q34.3
European Journal of Human Genetics (2010)