Abstract
Until now, over 30 loci have been identified by linkage analysis of affected families that segregate non-syndromic and dominantly inherited forms of hearing impairment (DFNA). A German family with a non-syndromic progressive hearing impairment transmitted in autosomal dominant mode was linked to 19q13.3-q13.4 by a genome-wide scan. Due to the low lod-score (1.85 at θ=0.05) for APOC2-locus we extended the fine mapping attempt with further markers in the same chromosomal region. This resulted in significant evidence for linkage to the markers D19S246 and D19S553 (two-point lod-score of 4.05 and 3.55 at θ=0.0) and a candidate critical region of 14 cM between markers D19S412 and D19S571. This region shows partial overlap with the previously reported DFNA4 critical region. The human gene BAX is orthologous to the rodent Bcl2-related apoptosis gene that is temporally expressed during the postnatal period in the developing inner ear of the mouse. BAX, mapping at a distance of no more than 0.73 cM distally to marker D19S553 appeared a likely candidate in our pedigree but genomic sequencing of coding regions and exon/intron boundaries excluded disease-related mutations. However, additional ESTs in the same region remain to be tested.
Similar content being viewed by others
Introduction
Hereditary hearing impairment is the most frequent sensorineural deficit in humans.1 It shows extreme genetic heterogeneity and phenotypic variability. The incidence of the prelingual hearing impairment is 1 of 1000 newborns and an equal number will progressively lose their ability to hear during their life.2 It has been assumed that in developed countries more than 60% of all cases are caused genetically.1 The non-syndromic forms of hearing impairment are usually monogenic and the mode of transmission is autosomal recessive in about 85% of cases, autosomal dominant in 15%, X-linked in 1–3% and mitochondrial in rare cases.1,3,4,5 Up to 200 genes are estimated either to be involved in the structural development of inner ear or to play a critical role in signal transduction.6 Through application of positional cloning strategies in affected families, more than 60 genomic regions co-segregating with non-syndromic hearing impairment have been identified in recent years (http://hgins.uia.ac.be/dnalab/hhh/). Until now, only 18 genes were cloned and studied at a molecular level.7 Eleven genes belong to the non-syndromic autosomal dominant segregating group of hearing loss. As expected, they participate in the development of the structure of inner ear (DFNA1; DFNA8/12; DFNA9; DFNA11; DFNA15) or play an important role in signal transduction and cell to cell communications (DFNA2; DFNA3). The function of DFNA5-gene is still unknown.8 Chen et al.9 reported identification of the DFNA4-locus on chromosome 19q13 and suggested DM-kinase as a possible candidate gene. In the present study we describe a German family with non-syndromic progressive hereditary hearing impairment showing an autosomal dominant mode of transmission linked to the chromosomal segment 19q13.3-q13.4. The critical region in our family overlaps only in part with the DFNA4 critical region and several ESTs require testing as candidate genes.
Materials and methods
Clinical data of the family Leipzig 9
All enrolled members of the family ‘Leipzig 9’ agreed to participate in the study. After obtaining informed consent, a general and otolaryngological examination was performed. History and physical examination was tailored to exclude syndromic forms of hearing impairment. The hearing status was evaluated with pure tone audiometry in the field with a portable Hortmann audiometer (Esslingen, Germany). In addition to pure tone audiometry, the subjects underwent transient evoked otoacoustic emission (TEOAE) testing to evaluate outer hair cell function. Children under the age of 10 underwent TEOAE testing and auditory brainstem response testing to supplement the pure tone hearing assessment. In addition, caloric vestibular testing was performed on affected members.
DNA analyses with microsatellite markers
Genomic DNA was prepared from peripheral blood lymphocytes according to Miller et al.10 Microsatellite polymorphisms were amplified by PCR with fluorescently labelled markers in a PTC-200 thermocycler (MJ Research). PCR conditions and details for all these markers can be found in the Genome Database (http://gdbwww.gdb.org). Fluorescent PCR products were detected by use of an ABI 377 DNA Sequencer (Applied Biosystems). Sizes of marker alleles were defined by use of GENESCAN software, version 3.1 (Applied Biosystems). Analysed data were imported to the GENOTYPER software package, version 2.0 (Applied Biosystems), to make a final table of estimated alleles. For a verification of segregation of estimated alleles in the family (allele-calling procedure), GENOTYPER-tables were imported to the LinkageDesigner software,7 to make a final table of genotypes, that were compatible for calculation of two point linkage analyses in LINKAGE software package ver. 5.1.11 All microsatellite markers used are listed in the Genethon map12 and/or the Genome Database (www.gdb.gov). In some cases (D19S571, D19S416, D19S418), the order of the markers differed in the different maps.
BAX sequencing
For PCR amplifications, a set of five primer pairs described by Yamamoto et al.13 allowed synthesis of the entire coding sequence of BAX gene. All reactions were performed in PTC-200 thermocycler (MJ Research) with DNA-Taq-Polymease from Gibco-BRL. The products were purified using the QIAquick PCR purification kit (Qiagen Hilden Germany) for direct sequencing of both strands with the same PCR-primers using the ABI PRISMTM Bigdye cycle sequencing kit (Perkin-Elmer Corp.) in an ABI-377 sequencing device.
Linkage analysis
Standard two point linkage analysis was performed by means of the LINKAGE programs11 assuming a fully penetrant autosomal dominant mode of inheritance with no phenocopies, disease allele frequency of 0.001, and equal marker allele frequencies.
Results
Linkage analysis and key recombinations
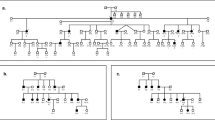
In a clinically well defined family ‘Leipzig 9’14 a whole-genome scan resulted in a positive but low lod-score for APOC2 (Table 1); thus seven new polymorphic microsatellite markers (D19S414, D19S425, D19S220, D19S412, D19S246, D19S553, D19S571) within a genetic distance of approximately 15 cM on either side of APOC2 in 19q13.2-q13.4 were chosen for further tests. D19S425 was originally used in the genome scan and six markers (see Table 1) were informative in our family. Maximum lod-scores of 4.05 and 3.55 were obtained at recombination fraction (θ)=0.0 from markers D19S246 and D19S553 respectively (Table 1). Key recombinations in the family were identified which reduced the span of the disease-specific haplotype to the 14 cM interval between markers D19S412 and D19S571 (Figure 1). This interval overlaps in part with the candidate region for DFNA4.9 In particular the 8 cM region between markers D19S412 and D19S246 is included in both our and DFNA4 candidate regions (Figure 2).
Schematic diagram of the investigated region between markers D19S414 and D19S571. The interval described by Chen et al.9 for DFNA4 with its candidate gene DMPK (lod-score for ApoC2 4.21) overlaps with the interval in family Leipzig 9 (lod score for D19S246 4.05 see Table 1). BAX was mapped distally to D19S553. Map positions are according to chromosome 19 databases http://cedar.genetics.soton.ac.uk/pub/chrom19/gmap and http:/www.ncbi.nlm.nih.gov/ (NT011190, HS chrom. 19 working draft sequence segment). Cochlea specific ESTs N64074, H88778, and H88366, located in the Leipzig 9 interval, originate from (http:/hearing.bwh.harvard.edu/cochlearcdnalibrary.htm).
D19S553 linked to BCL2-related apoptosis accelerating gene ‘BAX’
In order to screen for candidate genes and/or ESTs in our candidate critical region we searched the database of the British Medical Research Council (http://www.ihr.mrc.ac.uk/hereditary/genetable/index.html) as well as the Human cochlear cDNA library and EST-database (http://hearing.bwh.harvard.edu/chromosome19.htm). A member of the BCL2-apoptosis gene family, ‘BAX’ (BCL2-related X-protein), as well as ESTs: H88778, H88366 and N64074 map in our disease specific interval between D19S412 and D19S571 (Figure 2). Marker D19S553 is located 0.73 cM proximally to the locus of BAX.
No evidence for BAX mutations linked to the disease phenotype
Sequencing of the genomic regions and the exon/intron boundaries resulted in a regular BAX sequence in patients III-5 and V-2 and unaffected family members IV-3 and IV-6 from the pedigree as well as two unaffected and unrelated controls, therefore, very likely excluding this gene as a candidate for hearing impairment linked to 19q13.3-q13.4. A comparison of the 3′-end sequence of BAX with sequences of three ESTs (H88778, H88366 and N64074), located in the same region of disease specific haplotype, excluded any of them to represent BAX in cochlea specific cDNA-library.
Discussion
Chen et al.9 genetically analysed a progressive non-syndromic hearing loss in an American family and found a linkage with markers D19S425 and APOC2 located on chromosome 19q13 leading to definition of this locus as DFNA4. The family ‘Leipzig 9’ is to our knowledge the second family with non-syndromic hearing impairment linked to the chromosomal segment 19q13. Affected individuals show a progressive sensorineural hearing impairment that begins in the first to the second decade and leads to severe to profound hearing loss in the fourth decade of their lives. However, the disease-specific haplotype in the family ‘Leipzig 9’ with the highest lod-score of 4.05 and 3.55 at markers D19S246 and D19S553 shows only partial overlap to the critical region identified in the American family (see Figure 2).The candidate region in the ‘Leipzig 9’ family is defined at each side by recombination in individual III-2, who only shares alleles at two markers (D19S246 and D19S553) with his affected half-sister III-5 and cousin III-10. Possible explanations of our findings include the following.
First, it is possible that the same gene located in the interval between markers D19S412 and D19S246 is responsible for hearing impairment in both ours and the American family. Another explanation is that the hearing defect in the German family ‘Leipzig 9’ is caused by a mutation in a gene of a completely different function than the DFNA4-gene in the American family. Three candidate ESTs (N66534, N64583, H88351) in the subtracted cDNA-library are assigned to the affected DFNA-region described by Chen et al.9 Mutation in any of these three potential genes could be responsible for hearing deficit in the American family. Based on two reports about association of auditory abnormalities with myotonic dystrophy,15,16 Chen et al.9 assumed the possible involvement of the DM-kinase gene in developing of DFNA4, a fact yet needing proof of DM-kinase expression in the inner ear.
Linkage in the family ‘Leipzig 9’ seemed to offer an alternative candidate gene. An apoptosis accelerator protein, BAX ‘BCL2-related X-protein’, maps in 19q13.3-q13.4.17 Its specific expression in the developing inner ear of mouse18 suggested a similar importance in the human system. However, screening for mutations in BAX yielded no evidence for its fundamental role in developing hearing deficiency linked to 19q13.3-q13.4.
The cochlea-specific cDNA-library contains 3 ESTs: H88778, H88366 and N64074 mapping exactly in the disease specific haplotype of family ‘Leipzig 9’ described in this study (Figure 2). Analysis of the 3′-end sequence of BAX resulted in exclusion of any of these ESTs to represent BAX. The gene-rich region between the two flanking markers (D19S412 and D19S571) contains coding sequences for KCN2, ZNF160, FTL, CORD2, ZNF 83, FPRL, RRAS, SPIB, ZNF61, and ZNF50 (http://cedar.genetics.soton.ac.uk/pub/chrom19/gmap). Therefore, some of these ESTs (see Figure 2) could represent a potential gene underlying the hearing loss phenotype in the family ‘Leipzig 9’ and DFNA4.
References
Kalatzis V, Petit C . The fundamental and medical impacts of recent progress in research on hereditary hearing loss Hum Mol Genet 1998 7: 1589–1597
Skvorak AB, Weng Z, Yee AJ, Robertson NG, Morton CC . Human cochlear expressed sequence tags provide insight into cochlear gene expression and identify candidate genes for deafness Hum Mol Genet 1999 3: 439–452
Adato A, Raskin L, Petit C, Bonne-Tamir B . Deafness heterogeneity in a Druze isolate from the Middle East: novel OTOF and PDS mutations, low prevalence of GJB2 35delG mutation and indication for a new DFNB locus Eur J Hum Genet 2000 8: 437–442
Prezant TR, Agapian JV, Bohlman MC et al. Mitochondrial ribosomal RNA mutation associated with both antibiotic-induced and non-syndromic deafness Nature Genet 1993 4: 289–294
Torroni A, Cruciani F, Rengo C et al. The A1555G mutation in the 12S rRNA gene of human mtDNA: recurrent origins and founder events in families affected by sensorineural deafness Am J Hum Genet 1999 65: 1349–1358
Petit C . Genes responsible for human hereditary deafness: symphony of a thousand Nature Genet 1996 14: 385–391
VanCamp G, Smith RJH . Hereditary hearing loss homepage 2000 http:dnalab-www.uia.ac.be/dnalab/hhh
Van Laer L, Huizing EH, Verstreken M et al. Nonsyndromic hearing impairment is associated with a mutation in DFNA5 Nature Genet 1998 2: 194–197
Chen AH, Ni L, Fukushima K et al. Linkage of a gene for dominant non-syndromic deafness to chromosome 19 Hum Mol Genet 1995 4: 1073–1076
Miller SA, Dykes DD, Polesky HF . A simple salting out procedure for extracting DNA from human nucleated cells Nucl Acids Res 1985 16: 1215
Lathrop GM, Lalouel JM, Julier C, Ott J . Strategies for multilocus linkage analysis in humans Proc Natl Acad Sci USA 1984 81: 3443–3446
Dib C, Faure S, Fizames C et al. A comprehensive genetic map of the human genome based on 5,264 microsatellites Nature 1996 380: 152–154
Yamamoto H, Sawai H, Perucho M . Frameshift somatic mutations in gastrointestinal cancer of the microsatellite mutator phenotype Cancer Res 1997 57: 4420–4426
Oeken J, König S . Formen monosymptomatischer hereditärer Schallempfindungsschwerhörigkeiten und Taubheiten im Umkeis von Leipzig HNO 1993 41: 301–310
Huygen PLM, Verhagen WIM, Noten JFP . Auditory abnormalities, including ‘precocious prebycusis’ in myotonic dystrophy Audiology 1994 33: 73–84
Verhagen WIM, Bruggen PL, Huygen PLM . Oculomotor, auditory, and vestibular responses in myotonic dystrophy Arch Neurol 1992 49: 954–960
Apte SS, Mattei MG, Olson B . Mapping of the human BAX gene to chromosome 19q13.3-q13.4 and isolation of a novel alternatively spliced transcript, BAXδ Genomics 1995 26: 592–594
Ishii N, Wanaka A, Ohno K et al. Localization of bcl-2, bax, and bcl-x mRNAs in the developing inner ear of the mouse Brain Res 1996 726: 123–128
Acknowledgements
The authors are grateful to the family members for participating. This study was supported by the program fortüne (No. 591.0), BMBF (Fö 01KS9602) and the Interdisciplinary Center of Clinical Research (IZKF) Tübingen.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mirghomizadeh, F., Bardtke, B., Devoto, M. et al. Second family with hearing impairment linked to 19q13 and refined DFNA4 localisation. Eur J Hum Genet 10, 95–99 (2002). https://doi.org/10.1038/sj.ejhg.5200769
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5200769