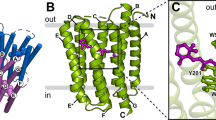
Abstract
Bacteriorhodopsin is the simplest known photon-driven proton pump1 and as such provides a model for the study of a basic function in bioenergetics. Its seven transmembrane helices2 encompass a proton translocation pathway containing the chromophore, a retinal molecule covalently bound to lysine 216 through a protonated Schiff base, and a series of proton donors and acceptors. Photoisomerization of the all-trans retinal to the 13-cis configuration initiates the vectorial translocation of a proton from the Schiff base, the primary proton donor, to the extracellular side, followed by reprotonation of the Schiff base from the cytoplasm. Here we describe the high-resolution X-ray structure of an early intermediate in the photocycle of bacteriorhodopsin, which is formed directly after photoexcitation. A key water molecule is dislocated, allowing the primary proton acceptor, Asp 85, to move. Movement of the main-chain Lys 216 locally disrupts the hydrogen-bonding network of helix G, facilitating structural changes later in the photocycle.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Oesterhelt,D. & Stoeckenius,W. Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nature New Biol. 233, 149–152 (1971).
Henderson,R. & Unwin,P. N. T. Three dimensional model of purple membrane obtained by electron microscopy. Nature 257, 28–32 (1975).
Landau,E. M. & Rosenbusch,J. P. Lipidic cubic phases: A novel concept for the crystallization of membrane proteins. Proc. Natl Acad. Sci. USA 93, 14532–14535 (1996).
Rummel,G. et al. Lipidic cubic phases: New matrices for the three dimensional crystallization of membrane proteins. J. Struct. Biol. 121, 82–91 (1998).
Schlichting,I., Berendzen,J., Phillips,G. N. Jr & Sweet,R. M. Crystal structure of photolysed carbonmonoxy-myoglobin. Nature 371, 808–812 (1994).
Genick,U. K., Soltis,S. M., Kuhn,P., Canestrelli,I. L. & Getzoff,E. D. Structure at 0.85 Å resolution of an early protein photocycle intermediate. Nature 392, 206–209 (1998).
Hadfield,A. & Hajdu,J. A fast and portable microspectrophotometer for protein crystallography. J. Appl. Crystallogr. 26, 839–842 (1993).
Doig,S. J., Reid,P. J. & Mathies,R. A. Picosecond time-resolved resonance Raman spectroscopy of bacteriorhodopsin's J, K and KL intermediates. J. Phys. Chem. 95, 6372–6379 (1991).
Xie,A. Quantum efficiencies of bacteriorhodopsin photochemical reactions. Biophys. J. 58, 1127–1132 (1990).
Bullough,P. A. & Henderson,R. The projection structure of the low temperature K intermediate of the bacteriorhodopsin photocycle determined by electron diffraction. J. Mol. Biol. 286, 1663–1671 (1999).
Braiman,M. & Mathies,R. Resonance Raman spectra of bacteriorhodopsin's primary photoproduct: Evidence for a distorted 13-cis retinal chromophore. Proc. Natl Acad. Sci. USA 79, 403–407 (1982).
Song,Q., Harms,G. S., Wan,C. & Johnson,C. K. Reorientations in the bacteriorhodopsin photocycle. Biochemistry 33, 14026–14033 (1994).
Takei,H., Gat,Y., Rothman,Z., Lewis,A. & Sheves,M. Active site lysine backbone undergoes conformational changes in the bacteriorhodopsin photocycle. J. Biol. Chem. 269, 7387–7389 (1994).
Luecke,H., Richter,H. T. & Lanyi,J. K. Proton transfer pathways in bacteriorhodopsin at 2.3 angstrom resolution. Science 280, 1934–1937 (1998).
Belrhali,H. et al. Protein, lipid and water organization in bacteriorhodopsin crystals: a molecular view of the purple membrane at 1.9 Å resolution. Structure 7, 909–917 (1999).
Gat,Y. & Sheves,M. A mechanism for controlling the pKa of the retinal protonated Schiff base in retinal proteins. A study with model compounds. J. Am. Chem. Soc. 115, 3772–3773 (1993).
Genick,U. K. et al. Structure of a protein photocycle intermediate by millisecond time-resolved crystallography. Science 275, 1471–1475 (1997).
Dencher,N. A., Dresselhaus,D., Zaccai,G. & Büldt,G. Structural changes in bacteriorhodopsin during proton translocation revealed by neutron diffraction. Proc. Natl Acad. Sci. USA 86, 7876–7879 (1989).
Koch,M. H. J. et al. Time-resolved X-ray diffraction study of structural changes associated with the photocycle of bacteriorhodopsin. EMBO J. 10, 521–526 (1991).
Nakasako,M., Kataoka,M., Amemiva,Y. & Tokunaga,F. Crystallographic characterization by X-ray diffraction of the M-intermediate from the photocycle of bacteriorhodopsin at room temperature. FEBS Lett. 292, 73–75 (1991).
Subramaniam,S., Gerstein,M., Oesterhelt,D. & Henderson,R. Electron diffraction analysis of structural changes in the photocycle of bacteriorhodopsin. EMBO J. 12, 1–8 (1993).
Vonck,J. A three-dimensional difference map of the N-intermediate in the bacteriorhodopsin photo-cycle: part of the F helix tilts in the M to N transition. Biochemistry 35, 5870–5878 (1996).
Nollert,P. & Landau,E. M. Enzymic release of crystals from lipidic cubic phases. Biochem. Soc. Trans. 26, 709–713 (1998).
Otwinowski,Z. in Data Collection and Processing (eds Sawyer, L., Isaacs, N. W. & Bailey, S.) DL/SCI/R34, 55–62 (Daresbury Laboratory, Warrington, 1993).
Collaborative Computational Project nr4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994).
Yeates,T. O. Detecting and overcoming crystal twinning. Methods Enzymol. 276, 344–358 (1997).
Valegård,K. et al. Structure of a cephalosporin synthase. Nature 394, 805–809 (1998).
Ursby,T. & Bourgeois,D. Improved estimation of structure-factor difference amplitudes from poorly accurate data. Acta Crystallogr. A 53, 564–575 (1997).
Brünger,A. T. et al. Crystallography and NMR system: A new software system for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998).
Jones,T. A., Bergdoll,M. & Kjeldgaard,M. in Crystallographic and Modelling Methods in Molecular Design (eds Bugg, C. & Ealick, S.) 189–199 (Springer, New York, 1990).
Esnouf,R. M. An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J. Mol. Graph. 15, 133–138 (1997).
Merritt,E. A. & Murphy,M. E. P. Raster3D version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr. D. 50, 869–873 (1994).
Acknowledgements
We thank D. Bourgeois, L.-O. Essen, R. Henderson, D. Oesterhelt, J. P. Rosenbusch, I. Schlichting and S. Subramaniam for discussions; W. P. Burmeister, A. Hardmeyer, T. Taylor and R. Wouts for experimental contributions; and G. Büldt for providing purple membrane. Support from the EU-BIOTECH, the Swedish Research Council NFR, the Institut Universitaire de France and the Swiss National Science Foundation's SPP BIOTECH is acknowledged. K.E. was an undergraduate student in the Molecular Biotechnology Programme of Uppsala University.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Edman, K., Nollert, P., Royant, A. et al. High-resolution X-ray structure of an early intermediate in the bacteriorhodopsin photocycle. Nature 401, 822–826 (1999). https://doi.org/10.1038/44623
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/44623
This article is cited by
-
Detailed analysis of distorted retinal and its interaction with surrounding residues in the K intermediate of bacteriorhodopsin
Communications Biology (2023)
-
True-atomic-resolution insights into the structure and functional role of linear chains and low-barrier hydrogen bonds in proteins
Nature Structural & Molecular Biology (2022)
-
Single molecule kinetics of bacteriorhodopsin by HS-AFM
Nature Communications (2021)
-
Improved production of bacteriorhodopsin from Halobacterium salinarum through direct amino acid supplement in the basal medium
Extremophiles (2019)
-
Tunable photocycle kinetics of a hybrid bacteriorhodopsin/quantum dot system
Nano Research (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.