Abstract
Skeletal muscle fibers form in overlapping, but distinct phases that depend on the generation of temporally different lineages of myogenic cells. During primary myogenesis (E10.5–E12.5 in the mouse), embryonic myoblasts fuse homotypically to generate primary fibers, whereas during later development (E14.5–E17.5), fetal myoblasts differentiate into secondary fibers. How these myogenic waves are regulated remains largely unknown. Studies have been hampered by the lack of markers which would distinguish embryonic from fetal myoblast populations. We show here that the homeobox gene Arx is strongly expressed in differentiating embryonic muscle, downstream of myogenic basic helix–loop–helix (bHLH) genes. Its expression progressively decreases during development. When overexpressed in the C2C12 myogenic cell line, Arx enhances differentiation. Accordingly, it stimulates the transcriptional activity from the Myogenin promoter and from multimerized E-boxes when co-expressed with MyoD and Mef2C in CH310T1/2. Furthermore, Arx co-immunoprecipitates with Mef2C, suggesting that it participates in the transcriptional regulatory network acting in embryonic muscle. Finally, embryonic myoblasts isolated from Arx-deficient embryos show a delayed differentiation in vivo together with an enhanced clonogenic capacity in vitro. We propose here that Arx acts as a novel positive regulator of embryonic myogenesis by synergizing with Mef2C and MyoD and by establishing an activating loop with Myogenin.
Similar content being viewed by others
Main
In amniotes, skeletal muscle differentiates in successive steps involving different type of myogenic progenitors.1 At about E11 in the mouse, during a phase usually referred to as primary myogenesis, embryonic myoblasts invade the limb buds and the pre-existing myotome, which is comprised of mononucleated differentiated myocytes, and fuse to form myotubes. These are the first multinucleated fibers that appear in the embryo and are termed ‘primary’ fibers. A second wave of myogenesis (secondary myogenesis) takes place between E14.5 and E17.5 and involves fusion of fetal myoblasts. At the end of gestation (after E16.5), emerging satellite cells can be identified as mononucleated cells located between the basal lamina and the fiber plasma membrane.2 Satellite cells are thought to be responsible for the postnatal growth and regeneration of the muscles. Work of several laboratories identified specific features of the different type of myoblasts. In particular, we recently reported that embryonic myoblasts have a greater propensity to differentiate compared to their fetal counterparts, since they more readily express markers of differentiation, have reduced proliferation and clonogenic ability in vitro.3
Embryonic, fetal myoblasts and satellite cells derive from a Pax3/Pax7-positive population of myogenic progenitors resident in the dermomyotome.4, 5 During skeletal muscle development these mesoderm-derived progenitors generate myoblasts, which become postmitotic and then terminally differentiate into multinucleated myotubes. The muscle regulatory factors (MRFs) MyoD, Myf5, Mrf4 and Myogenin6 are key regulators of the myogenic program and when transfected in fibroblasts, can convert them to a myogenic fate. Myf5, MyoD and Mrf4 can each independently initiate the myogenic program and thus act as myogenic ‘determination’ genes during primary myogenesis, whereas only Myf5 and MyoD can fulfill this role during secondary myogenesis.7 Conversely, Myogenin operates downstream of Myf5, MyoD and Mrf4.8
MRFs bind to E-box consensus sequences in the control regions of muscle-specific genes.6 Since all MRFs bind E-box motifs, additional transcription factors are required to regulate MRF specificity for particular target genes and to modulate their action. One such factor is MEF2 (Mef2A, Mef2B, Mef2C and Mef2D) that directly interacts with MRFs and synergizes with them.9 In addition to MEF2, a certain number of genes can modulate the action of the MRFs and thus positively or negatively regulate muscle differentiation.10
In this work, we investigated the role of the X-linked11 transcription factor Arx (Aristaless-related homeobox gene) during skeletal muscle development. Arx has been shown to be important for testes,12 central nervous system12 and pancreas development.13 Here, we show that Arx is predominantly expressed in embryonic myoblasts and primary fibers. Arx acts as positive modulator of myogenesis by synergizing with MyoD and Mef2C and establishing an activating loop with Myogenin. We therefore identify Arx as a novel positive regulator of embryonic myoblast differentiation. We propose that Arx is, at least in part, responsible for the accelerated differentiation of embryonic myoblasts.
Results
Developmental regulation of Arx expression in skeletal muscle
Quantitative PCR was performed on purified embryonic, fetal and postnatal myoblast cultures (Figure 1a). Arx expression was significantly higher in undifferentiated embryonic myoblasts compared to fetal and postnatal myoblasts (45.2- and 12.7-fold, respectively). Arx expression remained constant during differentiation of the embryonic population. In contrast, an increase in Arx transcription was observed during differentiation of the fetal population although the levels of Arx mRNA remained considerably lower than that in the embryonic population. Arx expression was also quantified on RNA from limbs and somites collected from E11.5 embryos and from fetal (E16.5) and adult limb muscles. As shown in Figure 1b, Arx expression markedly decreased during development.
Arx expression decreases during development. (a) Real-time PCR of Arx expression on embryonic myoblasts freshly isolated (column 1) or after 3 days in vitro (column 2); on fetal myoblasts freshly isolated (column 3), after 3 (column 4) or 6 days (column 5) in vitro; and adult myoblasts (derived from satellite cells) grown under proliferation-inducing conditions (column 6). (b) Arx expression assayed by real-time PCR on E11.5 embryos, E16.5 and adult limb muscles. Results have been normalized according to the expression of the muscle-specific gene α-actin; similar results were obtained when Mrf4 and Titin were used as normalizers
Arx expression in vivo was also investigated by whole-mount in situ hybridization (ISH). At the onset of primary myogenesis (E10–10.5), Arx was expressed in somites in a cranial-caudal gradient, both in the epaxial (dorsal) and hypaxial (ventral) domains of the myotomes. Expression was also detected in the first branchial arch, pancreas, neural tube and telencephalon (Figure 2a; data not shown). Significantly, Arx expression was not observed in newly formed myotomes at E9.5 (data not shown).14 At E11.5, Arx expression was apparent in the forming muscle masses of the limbs (Figure 2b) and at E13 in the intercostal, branchial and extraocular muscles (Figure 2d).
Arx is predominantly expressed during primary myogenesis. Whole-mount ISH of Arx in E10.5 (a), E11.5 (b) and E13 (d) wt mouse embryos. (c, c′) Co-immunohistochemistry on sections of lumbosacral somites of a E11.5 wt embryo (dotted line in b), using anti-Arx (red), anti-MyHC (green) antibodies and DAPI to stain the nuclei. X-Gal staining of E18.5 ArxnlacZ fetus (e) and diaphragms of ArxnlacZ (f) and Tg:3F-nlacZ-2E (f′) E18.5 fetuses. (g, g′) Co-immunohistochemistry on transverse sections of hind limbs of E16.5 ArxnlacZ fetuses, using anti-β-galactosidase (red), anti-MyHC (green) antibodies and Hoechst to stain the nuclei. Note that nuclear lacZ is expressed in a subset of fibers with large diameter (arrows) and is almost absent in surrounding small fibers (*). X-Gal staining of hind limb (h) and intercostal (h′) muscles of newborn (P7) ArxnlacZ mice and of soleus muscle (i, magnified in i′) and diaphragm (k) of adult Arx+/nlacZ mice. X-Gal (j) and hematoxylin and eosin (j′) staining of serial transverse sections of soleus muscle of adult Arx+/nlacZ mice revealed the expression of the reporter gene in spindle muscle fibers. Adult muscle could only be analyzed from Arx+/nlacZ mice, because mutant ArxnlacZ males die perinatally.13 Bar, 50 μm. BA, branchial arches; T, telencephalic vesicle; S, somites; first C1, cranial somite; T1, first thoracic somite; FL, forelimb; HL, hind limb; NT, neural tube; TA, tibialis anterior; Qu, quadriceps; G, gastrocnemius; IM, intercostal muscles; TM, temporalis muscle; BM, branchial muscles; R, ribs
Immunostaining showed co-localization of Arx protein with myosin heavy chains (MyHCs) in the myotome (Figure 2c, c′). In contrast, Arx was not detected in fetal or adult muscle, probably reflecting a low level of the protein (data not shown). However, a more sensitive analysis using a knock-in mouse-expressing nuclear β-galactosidase under the control of Arx regulatory sequences13 also revealed expression during later development. At fetal stages, X-Gal-positive nuclei were detected in all skeletal muscles (Figure 2e). These nuclei represent only a small fraction of the total number of muscle fiber nuclei as evidenced from a comparison of the staining in the ArxnlacZ with Tg:3F-nlacZ-2E fetuses (Figure 2f, f′). Interestingly, large primary fibers and smaller secondary fibers15 can be distinguished by the selective expression of Arx in the former, which presumably derive in large part from embryonic myoblasts (Figure 2g, g′).
After birth, a marked decrease in Arx expression, as assessed by X-Gal staining, was observed (Figure 2h, h′) and in adult muscle, expression was mainly restricted to a subset of fibers with the typical morphology of spindle fibers16 (Figure 2i, i′, j, j′). In contrast to the limb muscles, the diaphragm maintained a relatively wide Arx expression (Figure 2k). Collectively, these data suggest that Arx is predominantly expressed by embryonic myoblasts and their derivatives (primary fibers). Arx progressively decreases during development, remaining restricted in the adult to a subset of muscles (e.g., diaphragm) and specific fiber types (spindle fibers).
Arx expression depends upon the myogenic program activated by the myogenic determination genes MyoD, Myf5 and Mrf4 in the trunk, but not in the head.
To test whether Arx expression initiates after the acquisition of muscle identity, ISH was carried out on mutant embryos in which the expression of MyoD, Myf5 and Mrf4 is abolished (Myf5nlacZ/nlacZ(Mrf4):MyoD−/−). In these mutants, which completely lack skeletal muscle,7, 17 Arx expression was undetected in the somitic compartment, but was maintained in the pancreas and central nervous system (Figure 3b and d). Interestingly, Arx expression was maintained in cells colonizing the mesenchyme of the first branchial arch (Figure 3a and b, arrows), although starting from E11.5 the area of expression was reduced (Figure 3d, d′) in comparison with wild-type (wt) littermates (Figure 3c, c′).
ARX expression depends on the myogenic program activated by the MyoD, Myf5 and Mrf4 in the trunk. Whole-mount ISH of Arx in E10.5 (a, b) or E11.5 (c, c′, d, d′) wt (a, c, magnified in c′) and Myf5nLacZ/nLacZ(MRF4):MyoD−/− (b, d, magnified in d′) embryos. Note that in absence of all the three myogenic determination genes (Myf5, Mrf4 and MyoD) Arx expression in the muscle compartment is undetectable in the trunk, but is maintained in the first branchial arch (arrow). (e, e′) GFP-expressing cells are separated in two-dimensional dot plot of GFP (abscissa) and SSC (ordinate) from E11.5 Myf5GFP−P/+ (e) and Pax3GFP/+ (e′) embryos. (f) Arx expression was evaluated by real-time PCR on the GFP+ population isolated by FACS from Pax3GFP/+ (column 1) and Myf5GFP−P/+ (column 2). (g) Schematic representation showing the expression of Pax3, MRFs and Arx during the myogenic lineage progression at embryonic stage. An indicative quantification of the cells expressing Pax3 and Myf5 is indicated. T, telencephalic vesicles; P, pancreas; NT, neural tube
This observation suggests that Arx expression, at least in the trunk, is downstream of the myogenic program activated by the MRFs in the Pax3+/Pax7+ progenitors4, 5 (Figure 3g). Confirmation of this was obtained by the quantification of Arx transcripts in GFP+ cells isolated by fluorescence-activated cell sorter (FACS) from Myf5GFP−P/+ and Pax3GFP/+ embryos at E11.5. Pure populations of embryonic myoblasts were obtained from Myf5GFP−P/+ embryos, whereas a pool of Pax3+ progenitors and committed myoblasts was isolated from the trunk of Pax3GFP/+ embryos. Consequently, the number of GFP-expressing cells obtained from the Pax3GFP/+ embryos was significantly higher than the number of GFP+ cells sorted from the Myf5GFP−P/+ mice at the same stage (>10- fold; Figure 3e′, e). Quantitative PCR revealed a higher level of expression of Arx in the GFP+ cells isolated from the Myf5GFP−P/+ embryos compared with cells from the Pax3GFP/+ animals (9.5-fold; Figure 3f), consistent with the hypothesis that Arx is activated only after myogenic determination of the Pax3+ progenitors. Similarly to Arx, the myoblast marker desmin is also upregulated (3.4-fold) in the cells isolated from the Myf5GFP−P/+ embryos.
Arx positively modulates skeletal muscle differentiation
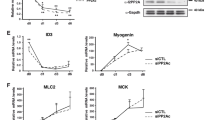
To understand the role of Arx, C2C12 myoblasts were transfected with vectors encoding ARX (in the sense or antisense orientation) and cytoplasmatic GFP, linked by an IRES element, thereby permitting the verification of efficient expression of the transfected plasmids by FACS analysis (Figure 4i). C2C12 were chosen since they do not express Arx before differentiation (data not shown). Only a negligible fraction of C2C12 transfected with the antisense cDNA expressed the differentiation markers Myogenin and MyHCs when cultured in 10% FCS, and this level was similar to the untransfected cells (2.7±1.3 versus 2.3±1.1% and 1.8±1.3 versus 1.8±0.4% for Myogenin and MyHCs, respectively). In contrast overexpression of human ARX (hARX) in C2C12 cells induced Myogenin and MyHCs in higher proportion (9.8±2 and 8.7±2.1%, respectively) (Figure 4a, a′, b, e, e′, f, j, k). The apparent increase in the differentiation of the cells overexpressing hARX was accompanied by increased levels of MLC1/3F and Myogenin, as determined by PCR and western blot (Figure 4l and m). Moreover, the hARX overexpressing cells showed reduced BrdU incorporation (32.2±4.7 versus 42.4±2.8% in the control cells), consistent with the observed enhanced differentiation state. After 4 days in low serum, the percentage of terminally differentiated (MyHCs+) cells was similar for both hARX overexpressing and control lines (Figure 4c, d, g, h). However, the hARX overexpressing myotubes were often branched and not aligned as observed in the control cultures (Figure 4c, d, g, h). Together, these observations suggest that Arx is able to positively modulate differentiation probably by accelerating the early stages of this process.
Enhanced differentiation in C2C12 stably transfected with a hARX. C2C12 were transfected with a vector containing hARX cDNA in antisense (control) or sense orientation under the control of CMV promoter. hARX-overexpressing (a, a′, b, c, d) and control (e, e′, f, g, h) C2C12 were grown in 10% FCS (proliferation medium) for 1 day (a, a′, b, e, e′, f) or in 2% FCS (differentiation medium) for 4 days (c, d, g, h). Immunostaining with anti-Myogenin (white; nuclear; a′, e′) and anti-MyHC (red; b, d, f, h) antibodies was performed. DAPI was used to stain the nuclei. Bar, 100 μm. A cytoplasmic GFP coding sequence was connected to the hARX cDNA by an IRES sequence. Stably transfected cells efficiently express GFP as assayed by FACS analysis (i). The number of Myogenin+ (j) and MyHC+ (k) cells was quantified on cells grown in proliferation medium and expressed as percentage of total cell number. Bars represent means of three experiments±S.D. *P<0.05. (l) Semiquantitative PCR analysis of Myogenin, Myosin light chain 1/3F (Myl1), hARX (sense) and Gapdh on hARX-expressing and control C2C12 grown in proliferation or differentiation medium. (m) Western blot illustrating upregulation of Myogenin protein in hARX-expressing C2C12 grown in 10% FBS. β-Tubulin was chosen for normalization and CH310T1/2 fibroblasts were used as negative control
Arx mutant embryos display delayed myogenesis
To investigate whether Arx is necessary for normal myogenic differentiation, we studied Arx-null mice. These mice die shortly after birth and display retarded growth, probably due to an abnormal pancreas.13 At the end of gestation (E19.5), no overt anomalies were observed in the growing muscle masses of mutant mice (data not shown) and adult heterozygous Arx+/nlacZ mice appeared normal, although some were obese. Nevertheless, at E11.5, MyHC+ muscles in the limb anlagen appeared smaller in the mutant embryos (Figure 5a and b); consistently, the pattern of expression of Myogenin (Figure 5c, c′, d, d′) was slightly delayed in most caudal somites. Moreover, myogenic clones from Arx mutant embryos were approximately twice larger than clones from wt littermates (Figure 5e). An ANOVA comparison of clone sizes confirms that the sizes are significantly different (P<0.01). These observations are consistent with Arx being a positive modulator of differentiation.
Embryonic myoblasts collected from ArxnlacZ mutant mice present a delayed differentiation. Immunostaining with anti-MyHC antibodies on transverse sections of E11.5 wt (a) and Arx-null mutant (b) littermates. DAPI was used to stain the nuclei. Whole-mount ISH of Myogenin in E11.5 wt (c, c′) and Arx mutant (d, d′) littermates. NT, neural tube; M, myotome; FL, forelimb; HL, hindlimb; C1, T1, L1, S1, Co2 respectively first cranial, thoracic, lumbar, sacral and second caudal somite. Note that third, fourth, fifth and sixth caudal somites express Myogenin only in wt embryos (arrows). (e) Cells collected from single E11.5 wt (n=7), heterozygous (n=1) and Arx mutant (n=6) embryos were plated at clonal density and colonies of myogenic cells were recognized by staining with anti-MyoD antibodies. For each embryo, the number of cells (MyoD+) was counted in at least 50 myogenic clones and expressed as average number of cells per myogenic clone. Three different litters were independently analyzed (exp. 1, 2 and 3). Note that the myogenic clones of Arx-deficient embryos consistently contain more cells in comparison to their wt littermates
Arx synergizes with MyoD and Mef2C in the activation of muscle-specific transcription
We then tested the ability of Arx to activate the Myogenin promoter in a chloramphenicol acetyltransferase (CAT) assay performed on CH310T1/2 that do not normally express the gene. When CH310T1/2 were transfected with an ARX-expressing vector, myogenic conversion was not observed (data not shown). However, when transfected with combinations of vectors encoding ARX or MyoD and a CAT reporter vector carrying the 5′ regulatory region of mouse Myogenin, a synergistic effect of Arx and MyoD was observed with a threefold increase in reporter activity (Figure 6a). Since Mef2C is known to modulate positively the myogenic conversion induced by MRFs in transfected fibroblast,19 we tested the action of Arx in the presence of both exogenous Mef2C and Myod. The results of this experiment showed an ulterior increase (fivefold increase over MyoD alone; Figure 6a). Thus, Arx synergizes with MyoD in the activation of the Myogenin promoter and the efficiency of this activation is enhanced by increased levels of Mef2C; the ARX vector alone or co-transfected with a Mef2C-expressing plasmid did not significantly increase CAT expression over the basal level. The same assay has also been performed with a reporter construct containing a multimerized E-box,20 further confirming the strong synergism (4.9-fold over MyoD alone) between the three factors (Figure 6b). Under these experimental conditions, the activation of the reporter vector was only barely enhanced when alternatively Arx or Mef2C were co-transfected with MyoD (Figure 6b).
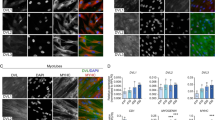
Arx positively modulates the transcription from the Myogenin promoter and from an E-box-containing reporter vector, by participating in a complex with Mef2C and MyoD. Relative CAT activities, corrected for differences in transfection efficiencies, were determined for extracts from CH310T1/2 fibroblasts transfected alternatively with the reporter vectors pMyo-1565CAT (myog) (a) or 4RCAT (4xE-box) (b). Values are expressed in arbitrary units relative to the CAT activity produced by the control plasmid, which was assigned a value of 1. The data are averages of at least three independent experiments performed in duplicate. Bars represent means±S.D. *P<0.05 versus MyoD and MyoD+Mef2C+ARX; ***P<0.001 versus MyoD and MyoD+ Mef2C. (c) The expression of Mef2C has been evaluated by real-time PCR on the embryonic population freshly isolated (column 1) or after 3 days in vitro (column 2) and on the fetal population freshly isolated (column 3) after 3 (column 4) or 6 days (column 5) in vitro and satellite cells (column 6). (d) C2C12 were transfected with a plasmid-expressing Myc-tagged GFP–ARX fusion protein and co-immunoprecipitated (IP) with anti-Myc (d′), anti-MyoD (d″) or anti-Mef2C (d″′) antibodies. Western blot showed that MyoD and Mef2C form a complex with ARX. Immunoprecipitation with rabbit anti-GFP antibodies was also performed and give similar results (data not shown). Lysates from untransfected CH310T1/2 and from transfected, but not immunoprecipitated C2C12 (Tot) have been used as negative and positive controls, respectively. (e) CH310T1/2 were transfected with Myc-ARX and pMef2C (e′); pEMSV-Myf3 (Myod1) and pMef2C (e″) or pEMSV-Myf3 and Myc-ARX (e″′ , e″″) plasmids and immunoprecipitated with anti-Myc (e′, e″″), anti-MyoD (e″, e″′) antibodies. Western blot indicated that Mef2c can interact with both ARX and MyoD, whereas MyoD and ARX likely do not bind directly to each other. Immunoprecipitations with anti-Mef2C antibodies confirmed the interaction of Mef2C with MyoD and ARX in CH310T1/2 (data not shown). Immunoprecipitations with non-programmed IgGs showed the specificity of the immunoprecipitations. The asterisks indicate two bands corresponding to hypo- (fast-migrating) and hyper- (slow-migrating) phosphorylated forms of MyoD18
Intriguingly, real-time PCR approach showed that Mef2C, like Arx, is more expressed in embryonic myogenic cells in comparison to their fetal counterparts, both before and after differentiation (5.8- and 2.9-fold, respectively; Figure 6c).
To investigate a possible physical interaction between the Arx , MyoD and Mef2C proteins, C2C12 were transfected with a vector expressing a Myc-tagged GFP–ARX fusion protein. The tagged protein was then immunoprecipitated with anti-Myc antibodies and the immunoprecipitate probed for MyoD on western blot. The results shown in Figure 6d′ reveal that the exogenous, tagged ARX co-immunoprecipitated with endogenous MyoD. Figure 6d′ shows the presence of Mef2C in the same immunoprecipitate, suggesting an interaction with Arx. The interaction between Arx, MyoD and Mef2C was further confirmed by the presence of Myc–GFP–hARX fusion protein in the immunoprecipitate after co-immunoprecipitation with anti-MyoD (Figure 6d″) or anti-Mef2C (Figure 6d″′) antibodies. Collectively, these data suggest that Arx, MyoD and Mef2C proteins are able to participate into the same complex and that Arx could act as co-activator during myogenic differentiation. Nevertheless, since a direct binding between MRFs and Mef2 proteins has been reported,19 it is possible that Arx directly associate either with MyoD or Mef2C and the other factor co-immunoprecipitate with Arx because the Mef2 proteins interact with MyoD. To address this point, CH310T1/2 (that do not autonomously express MyoD and Mef2C at variance with C2C12) were co-transfected with different combinations of two plasmids expressing alternatively MyoD, Mef2C or Myc-tagged ARX. These experiments confirmed the physical interaction between MyoD and Mef2C (Figure 6e″) and revealed that, under these conditions, Arx co-immunoprecipitate with Mef2C (Figure 6e′), but not with MyoD (Figure 6e″′, e″″). Taken together, these data suggest that Mef2C binds both to Arx and MyoD, whereas MyoD and Arx do not directly interact with each other.
Arx establishes a positive regulatory loop with Myogenin
MRFs are required for Arx expression in the embryo (shown above) and in CH310T1/2 (data not shown). However, in CH310T1/2, identification of the specific MRF that activates Arx is complicated by the ability to cross- and auto-activate expression from the endogenous loci.21 To test whether Arx expression could be induced independently by the different MRFs, mouse embryonic fibroblasts (MEFs) were isolated from Myf5nlacZ/nlacZ(Mrf4):MyoD−/− mice which lack Myf5, Mrf4 and MyoD. These cells were transfected with plasmids encoding MYF5, MyoD, MRF4 or Myogenin and the expression of Arx was assayed by semiquantitative RT-PCR (Figure 7a). Myogenin activated endogenous Arx expression in absence of the other MRFs. Since ARX can enhance activation of the Myogenin promoter induced by MyoD in CH310T1/2 (shown above), it is likely that Arx and Myogenin directly or indirectly activate each other's expression, suggesting the existence of a positive regulatory loop between these transcription factors. MRF4 and MYF5 also activated Arx expression in fibroblasts from Myf5nlacZ/nlacZ(Mrf4):MyoD−/− embryos, whereas MyoD did so only faintly. MyoD, Myf5 and Mrf4 are all able to cross-activate Myogenin. Nevertheless, it seems unlikely that the effects of the other MRFs on Arx transcription could be entirely mediated by Myogenin. In fact, when expressed in fibroblasts from Myf5nlacZ/nlacZ(Mrf4):MyoD−/− mice, either MRF4 or MYF5 activated Arx more efficiently than MyoD despite the lower activation of Myogenin (Figure 7a). The differential ability to induce Arx shown by the Myf5, Mrf4 and MyoD in vitro is consistent with their in vivo effect. In all mutant embryos considered (MyoD−/−, Myf5loxP/loxP Myf5loxP/loxPMyoD−/− and Myf5nlacZ/nlacZ(Mrf4)), Arx expression was reduced (Figure 7b, c, d, e, f); however, only when Myf5 and Mrf4 were switched off, the reduction was dramatic suggesting that both genes are required for full activation of Arx. Nevertheless, although strongly delayed, the appearance of Arx transcripts in the absence of Myf5 and Mrf4 (Figure 7f, arrows) indicates that MyoD and possibly Myogenin can eventually activate Arx.
Arx expression can be differentially influenced by different myogenic regulatory factors. (a) MEFs collected from E12.5 Myf5nlacZ/nlacZ(MRF4) embryos were transfected with plasmids expressing Myf5, MyoD, Mrf4 and Myogenin and grown for 48 h in medium containing 10% (L) or 2% (H) FCS. Note that Arx expression is induced only when cells were maintained in 2% FBS. pEMSV-scribe was used as negative control. Whole-mount ISH analysis of Arx in E12 wt (b), MyoD−/− (c), Myf5loxP/loxP (d), Myf5loxP/loxPMyoD−/− (e) and Myf5nlacZ/nlacZ(Mrf4) (f) embryos
Discussion
The data reported here show that Arx is expressed in mouse embryonic muscles; its expression is dependent upon the myogenic program activated by MyoD, Myf5 and Mrf4 in somitic muscles, but interestingly not in the first branchial arch. Although further studies are required to understand the fate that Arx-expressing cells in the branchial adopt in mice mutant for MyoD, Myf5 and Mrf4, this observation is in agreement with previous findings that reported differences in the head and somitic myogenic programs.7, 22
During later development, the expression of Arx progressively decreases. Our PCR data are consistent with the pattern of β-galactosidase expression observed in Arx knock-in mice and with a microarray analysis performed on purified embryonic and fetal myoblasts.3 In the adult Arx+/nlacZ mice, β-galactosidase expression was maintained in spindle fibers of appendicular muscles and in a large proportion of fibers in the diaphragm. Interestingly, it has been proposed that a subset of spindle fibers may originate from embryonic myoblasts.16 This is supported by the high expression of Arx in both embryonic myoblasts and spindle fibers.
Arx is strongly expressed in proliferating embryonic myoblasts but only weakly in fetal and adult myoblasts, suggesting that it may play different roles in the different myogenic populations. When hARX was overexpressed in C2C12, precocious expression of differentiation markers was observed in conditions that promote proliferation. This mimics the phenotype of embryonic myoblasts, which express high level of endogenous Arx and are particularly prone to differentiate.3 Conversely, embryonic myoblasts from Arx mutant mice, which do not express Arx, showed a delayed differentiation in vivo together with an enhanced ability to proliferate in vitro. It is likely that other proteins may cooperate with Arx to enhance the differentiation of embryonic myoblasts and it is conceivable that they may also partially compensate for the absence of Arx, which by itself does not prevent the formation of an apparently normal skeletal muscle at fetal stage. Other myogenic regulators such as Mef2C (see Figure 6c) are also more highly expressed in embryonic compared with fetal and adult myoblasts. Interestingly, neither Mef2C nor Arx are able to induce myogenic conversion in fibroblasts, but they both appear to be able to modulate positively the myogenic program in presence of MRFs (Lassar et al18 and present work). We show here that Mef2C physically interacts with both Arx and MyoD, and that in transfected CH310T1/2, a strong synergy occurs between the three factors in the activation of the transcription from Myogenin promoter and multimerized E-boxes containing reporter constructs, suggesting a cooperation in the induction of the myogenic program. Interestingly, previous studies demonstrated that E proteins are required for MyoD function and that the level of endogenous E proteins in CH310T1/2 is sufficient to support myogenic conversion.18 In contrast, when MyoD and Mef2 were tested for their ability to synergize on artificial reporters, they also required exogenous E12,19 suggesting that E proteins are limiting for activation of exogenous reporter genes that are rather present at high copy number. The data reported here indicate that forced expression of ARX can somehow substitute for the absence of additional (exogenous) E proteins. Taken together, these observations favor a model in which MyoD, Mef2C and Arx create a multiprotein complex that interacts with the transcriptional machinery more efficiently than any of the individual factors.
Although Arx contains a homeobox DNA-binding domain,23 it is currently unknown whether its interaction with the Myogenin promoter is direct, mediated or stabilized by other factors. Moreover, as Arx appears to interact physically with Mef2C, but not directly with MyoD, the ability shown by Arx to activate to a certain extent the Myogenin promoter in CH310T1/2 in absence of exogenous Mef2C suggests that Mef2C may be induced by MyoD24 or that other factors participate in the activation. Arx has been suggested to be a bifunctional transcription factor, capable of functioning either as a repressor25, 26 or an activator of transcription.27 The observations reported here suggest that in the context of embryonic myogenesis, Arx positively modulates transcription. The synergism between Arx, Mef2C and MyoD could result from a specific recognition event between the multiprotein complex and a component of the transcriptional machinery or it could result from a concerted reaction in which the assembly of the three factors stabilizes a transcriptional complex, similarly to what was reported for MyoD and Pbx, the latter containing a homeodomain like Arx.28
The data presented here also show that Arx and Myogenin can upregulate each other's expression. Similarly, Mef2 gene products also regulate Myogenin expression9, 29 and the Mef2 genes are known targets of the myogenic basic helix–loop–helix (bHLH)(24). It is therefore likely that Arx, probably by co-activating MyoD and Mef2C, enhances the transcriptional circuitry that regulates progression in the lineage to differentiation. In particular, the higher levels of Arx (and Mef2C) present in embryonic myoblasts may contribute to the enhanced differentiation ability that characterizes this myogenic lineage.
MyoD, Myf5, Mrf4 and Myogenin appear to play different roles in regulating Arx expression. Myogenin establishes a positive regulatory loop with Arx, which is independent of the other MRFs in MyoD/Myf5/Mrf4-null fibroblasts. Both in vitro and in vivo data suggest that the other MRFs can also activate the expression of Arx albeit with a different efficiency. Differential trans-activation of muscle-specific genes by different MRFs has already been reported.30 When fibroblasts were transfected with MRFs-encoding plasmids, Arx expression was only induced upon myogenic differentiation (i.e., grown in low serum). In this respect, fibroblasts converted to a myogenic fate by forced MRF expression differ from embryonic myoblasts which are the only cells expressing Arx during proliferation, suggesting the existence of additional factors specific to these cells. Microarray analysis showed that embryonic and fetal myoblasts express similar level of all four MRFs.3 Therefore, although Arx expression depends on Myf5, Mrf4 and MyoD, other still not identified proteins must be responsible for the enhanced expression of Arx in embryonic myoblasts.
In conclusion, the data presented are the first demonstration that a homeodomain gene differentially regulates embryonic versus fetal myoblast differentiation. Arx activity involves cooperation with MyoD and Mef2C, both of which are also highly expressed in embryonic myoblasts. Moreover, Arx establishes a positive regulatory loop with Myogenin that enhances the transcriptional circuitry regulating differentiation (Figure 8).
Genetic hierarchy governing embryonic skeletal muscle differentiation. Arx and Mef2C (in red) are predominantly expressed in embryonic myoblasts. Arx synergizes with Mef2C and MyoD. Arx and Myogenin participate in a positive regulatory loop in differentiating embryonic myoblasts. Arrows depict genetic relationships, and do not necessary imply direct interactions. For the purpose of simplification, cross-activation between MRFs and Mef2 proteins and Myogenin-independent activation of muscle genes by MyoD and Mrf431 are not represented
Materials and Methods
Cell isolation
Embryonic and fetal myoblasts were isolated respectively from E11.5 and E16.5 Myf5GFP−P/+ mice7 (after removal of the head, internal organs and skin). Tissues were dissociated with 0.15 mg/ml Collagenase Type V (Sigma, St. Louis, Mo, USA), 0.4 mg/ml Dispase (Gibco-Invitrogen, Carlsbad, CA, USA), 0.1 mg/ml DNase I (Roche, Mannheim, Germany). When GFP-expressing cells were isolated from Pax3GFP/+ 5 E11.5 embryos, the neural tube and dorsal root ganglia were removed. The lack of any neural crest contamination was confirmed by the absence of Sox10-positive cells in the sorted population (data not shown). GFP-expressing cells were sorted using a Vantage Sorter SE (BD, San Diego, CA, USA). Forward (FSC) and Side Scatter (SSC) parameters were used to gate out cell clumps and debris. Cells dissociated from wt littermates were used to set the gating to exclude green autofluorescence.
Satellite cells were isolated from 3- to 5-day-old mice as described previously.32 Cells were grown at clonal density in DMEM-high glucose supplemented with 20% fetal calf serum (FCS; BioWhittaker-Cambrex, Verviers, Belgium), 20 mM HEPES and 3% chicken embryo. Clones of spindle-shaped satellite cells were trypsinized in cloning cylinders (Sigma) and pooled together. The resulting satellite cell population was 95% pure as assayed by staining with anti-desmin antibody.
Cell culture
To assay colony-forming capacity, unsorted cell suspensions from dissociated wt or Arx-mutant13 E11.5 and E16.5 mice were plated at clonal density (approximately 200 cells/cm2) and cultured in DMEM-high glucose supplemented with 20% horse serum (BioWhittaker-Cambrex), 20 mM HEPES and 3% chicken embryo extract for 3 days. The myogenic colonies were identified with anti-MyoD antibody.
C2C12 myoblasts were plated at 20 000 cells/cm2 in DMEM-high glucose, supplemented with 10% FCS. The day after plating, cells were either processed as proliferating cultures, or the medium was replaced with DMEM-high glucose supplemented with 2% FCS to induce differentiation.
Immunofluorescent and β-galactosidase staining of cultured cell and tissue sections
Cell cultures and 8-μm cryostat sections were fixed in 4% paraformaldehyde for 10 min at 4°C. For β-galactosidase, staining sections were incubated at 37°C for 5 h in X-Gal solution (1 mg/ml 5-bromo-4-chloro-3-indolyl-β-D-galactosidase, 5 mM K4Fe(CN),6 5 mM K3Fe(CN),6 0.02% NP-40, 2 mM MgCl2 in PBS). Where necessary, adjacent sections were stained with hematoxylin and eosin. For evaluating the proliferative capacity, cells were incubated with 50 μm BrdU (Sigma) for 1 h, washed, fixed in a 95% ethanol/5% (v/v) acetic acid solution for 20 min and then treated with 1.5 M HCl for 10 min. Immunofluorescent staining was carried out using the following antibodies: rabbit polyclonal33 and mouse monoclonal (MF20) antibody to all sarcomeric MyCHs; mouse anti-BrdU (Amersham, Buckinghamshire, UK); mouse anti-Myogenin (F5D); rabbit anti-Arx,13 mouse anti-MyoD antibodies (clone 5.8A, DakoCytomation, Carpinteria, CA, USA), goat anti-β-galactosidase antibodies (Biogenesis, Poole, UK). Secondary antibodies used are Alexa Fluor 594-conjugated donkey anti-rabbit IgG, Alexa Fluor 546-conjugated donkey anti-goat IgG or Alexa Fluor 488-conjugated donkey anti-mouse IgG antibodies (Molecular Probes, Leiden, The Netherlands). DAPI or Hoechst were used to stain the nuclei.
Quantitative real-time polymerase chain reaction
Total cellular RNA was isolated from cell populations using RNeasy RNA isolation kit (Qiagen, Hilden, Germany). cDNAs were reverse transcribed from total RNA samples (100 ng/sample) using High Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA, USA). TaqMan PCR reactions were carried out from cDNA samples using custom TaqMan low-density arrays (Applied Biosystems) by means of ABI PRISM 7900 HT Sequence Detection Systems. Gene expression profiling was achieved using the Comparative CT method of relative quantification. Until otherwise is not stated the ribosomal RNA 18 s subunit was used as an internal standard.
Transfection of C2C12 and MEFs
A BglII/EcoRI fragment containing the full-length human ARX cDNA was cloned into in pIres2-EGFP (Clontech, Mountain View, CA, USA), cut either with BglII/EcoRI or BamHI/EcoRI to generate phARXiresEGFP sense and antisense, respectively. C2C12 myoblasts were transfected using Lipofectamine with Plus reagent (Invitrogen, Carlsbad, CA, USA). Cytoplasmatic GFP-expressing cells were purified by FACS the day after transfection and then cultured in DMEM supplemented with 10% FCS under selection with G418. Efficient expression of the transgene was verified in stably transfected cells by FACS analysis for GFP and in the case of hARX cloned in the sense orientation (hARX sense), also by RT-PCR using a primer located in the ARX coding sequence and another in the IRES: (5′-CGGCATTCGGCAGGCTCT-3′; 5′-AGGAACTGCTTCCTTCACGA-3′).
MEFs were isolated from Myf5nlacZ/nlacZ(Mrf4):MyoD−/− E12.5 embryos. Cells were grown for six passages in DMEM supplemented with 20% FCS and transiently transfected with Lipofectamine with Plus reagent (Invitrogen). For the transfection, vectors encoding rat Myogenin, mouse MyoD, human MYF5 or MRF4 under the control of the Moloney murine sarcoma virus long terminal repeat (MoMSV-LTR) or an empty vector (pEMSV α-scribe) were used (provided by Dr. M Grossi). After transfection, cells were maintained either in DMEM with 10% FCS or 2% FCS for 2 days before RNA extraction.
For semiquantitative PCR, cDNA was reverse transcribed from total RNA with ThermoScript RT-PCR (Invitrogen), using random hexamers according to the manufacturer's instruction. The primers used to amplify Myosin light polypeptide 1 (Myl1) were 5′-GATCACCTTAAGTCAGGT-3′; 5′-GCAAGGCTTCTACCTCTT-3′. Those used to amplify hMYF5 transcripts are 5′-GCCTTCTTCGTCCTGTGTATTAG-3′; 5′-TGAGAGAGCAGGTGGAGAACTAC-3′. The other primers used have been published.34, 35 All primers were designed to cross an intron/exon boundary. In the case of Arx amplification, 5% DMSO was added to the reaction mix.
CAT assay
CH310T1/2 fibroblasts were transiently transfected with Lipofectamine with Plus reagent (Invitrogen). Briefly, cells were seeded at 105 on 35 mm dishes and the following day, transfected alternatively with 0.5 μg of the reporter construct pMyo-1565CAT36 or 4RCAT (containing the tk minimal promoter and four copies of the right E-boxes from the MCK enhancer20) and 0.5 μg pCMV-βGal (Clontech) alone or together with various combinations of plasmids mixed as follows: 1 μg of pEMSV α-scribe+0.5 μg pires2EGFP (control); 0.5 μg of pEMSV-Myf3 (Myod1)+0.5 μg pires2EGFP+0.5 μg of pEMSV α-scribe; 0.5 μg phARXiresEGFPsense+1 μg of pEMSV α-scribe; 0.5 μg of pEMSV-Myf3+0.5 μg phARXiresEGFPsense+0.5 μg of pEMSV α-scribe; 0.5 μg of TA-Mef2C+ 0.5 μg pires2EGFP+0.5 μg of pEMSV α-scribe; 0.5 μg of TA-Mef2C+ 0.5 μg phARXiresEGFPsense+0.5 μg of pEMSV α-scribe and 0.5 μg of pEMSV-Myf3+0.5 μg pires2EGFP+0.5 μg of TA-Mef2C . TA-Mef2C was generated by inserting the murine Mef2C cDNA in the HindIII/XbaI sites of pFlag-CMV vector (Sigma). Cells were grown for 2 days in DMEM, 10% FCS and proteins were then extracted by repeated freezing-thawing. The levels of CAT protein were determined using an enzymatic immunoassay (CAT-ELISA/Roche). Cell extracts were normalized for protein concentration and CAT expression was further normalized to β-galactosidase activity.
Mutant animals and genotyping
The mutant and transgenic animals and genotyping primers used in this work were published: Pax3GFP,5 ArxnlacZ,13 Tg:3F-nlacZ-2E,37 MyoD−/−,38 Myf5GFP−P, Myf5loxP, Myf5nlacZ.7, 39 In Tg:3F-nlacZ-2E mice, β-galactosidase expression is restricted to differentiated skeletal and cardiac muscle.37 Myf5nlacZ/nlacZ mice have a loss of function of both Myf5 and Mrf4 at embryonic stages, whereas in Myf5loxP/loxP Mrf4 expression appears unaffected.7 Double mutant Myf5loxP/loxPMyoD−/− and Myf5nlacZ/nlacZ(Mrf4):MyoD−/− embryos were obtained by breeding double heterozygous mice. Embryonic day E0.5 corresponds to the morning of the vaginal plug. All experiments were performed under internal regulations for animal care and handlings.
Whole-mount ISH and X-Gal staining
Whole-mount ISH was performed according to standard protocols. RNA probes labeled with digoxigenin-UTP (Roche) were synthesized from linearized cDNA templates according to manufacturer's instructions. A fragment corresponding to the full-length mouse Arx cDNA34 and a 695-bp fragment corresponding to the 3′ end of murine Myogenin cDNA were used as riboprobes. For whole-mount X-Gal staining, embryos and isolated muscles were fixed overnight in 4% paraformaldehyde at 4°C and stained as described for the sections.
Immunoprecipitation and western blot
C2C12 myoblasts were transiently transfected with a vector encoding a Myc-tagged GFP–hARX fusion protein (Myc-ARX).26 CH310T1/2 fibroblasts were transiently transfected with various combinations of plasmids mixed as follows: 1 μg of Myc-ARX+ 1 μg of TA-Mef2C; 1 μg of pEMSV-Myf3 (Myod1)+1 μg of TA-Mef2C or 1 μg of pEMSV-Myf3+1 μg of Myc-ARX. The immunoprecipitation assay was performed as described previously40 by using mouse anti-Myc tag, (Roche), mouse anti-MyoD (Dako) or goat anti-Mef2C (Santa Cruz Biotechnology, Santa Cruz, CA, USA) antibodies. Immunoprecipitations with normal goat and mouse IgG (Santa Cruz) were used as controls.
For western blot analysis, immunoprecipitates and total extracts were resolved by SDS-PAGE on 10% polyacrylamide gels and then transferred onto nitrocellulose. After blocking in 5% milk in Tris-buffered saline (TBS)-T (TBS plus 0.02% Tween 20), filters were incubated with anti-MyoD, anti-Myogenin, mouse anti-Mef2C (Cell Signaling, Danvers, MA, USA) or mouse anti-tubulin (Covance, Princeton, NJ, USA) antibodies. After being washed, membranes were incubated with horseradish peroxidase-conjugated, species-specific secondary antibodies (Bio-Rad, Hercules, CA, USA) followed by enhanced chemiluminescence system (Amersham).
Abbreviations
- bHLH:
-
basic helix-loop-helix
- CAT:
-
chloramphenicol acetyltransferase
- FACS:
-
fluorescence-activated cell sorter
- ISH:
-
in situ hybridization
- MEF:
-
mouse embryonic fibroblast
- MRF:
-
muscle regulatory factor
- MyHC:
-
myosin heavy chain
- wt:
-
wild-type
References
Cossu G, Biressi S . Satellite cells, myoblasts and other occasional myogenic progenitors: possible origin, phenotypic features and role in muscle regeneration. Semin Cell Dev Biol 2005; 16: 623–631.
Mauro A . Satellite cell from skeletal muscle fibers. J Biophys Biochem Cytol 1961; 9: 493–495.
Biressi S, Tagliafico E, Lamorte G, Monteverde S, Tenedini E, Roncaglia E et al. Intrinsic phenotypic diversity of embryonic and fetal myoblasts is revealed by genome-wide gene expression analysis on purified cells. Dev biol 2007; 304: 633–651.
Kassar-Duchossoy L, Giacone E, Gayraud-Morel B, Jory A, Gomes D, Tajbakhsh S . Pax3/Pax7 mark a novel population of primitive myogenic cells during development. Genes Dev 2005; 19: 1426–1431.
Relaix F, Rocancourt D, Mansouri A, Buckingham M . A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature 2005; 435: 948–953.
Pownall M, Gustafsson M, Emerson CJ . Myogenic regulatory factors and the specification of muscle progenitors in vertebrate embryos. Annu Rev Cell Dev Biol 2002; 18: 747–783.
Kassar-Duchossoy L, Gayraud-Morel B, Gomes D, Rocancourt D, Buckingham M, Shinin V et al. Mrf4 determines skeletal muscle identity in Myf5:Myod double-mutant mice. Nature 2004; 431: 466–471.
Tajbakhsh S, Buckingham M . The birth of muscle progenitor cells in the mouse: spatiotemporal considerations. Curr Top Dev Biol 2000; 48: 225–268.
Black B, Olson E . Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu Rev Cell Dev Biol 1998; 14: 167–196.
Sabourin L, Rudnicki M . The molecular regulation of myogenesis. Clin Genet 2000; 57: 16–25.
Ohira R, Zhang Y, Guo W, Dipple K, Shih S, Doerr J et al. Human ARX gene: genomic characterization and expression. Mol Genet Metab 2002; 77: 179–188.
Kitamura K, Yanazawa M, Sugiyama N, Miura H, Iizuka-Kogo A, Kusaka M et al. Mutation of ARX causes abnormal development of forebrain and testes in mice and X-linked lissencephaly with abnormal genitalia in humans. Nat Genet 2002; 32: 359–369.
Collombat P, Mansouri A, Hecksher-Sorensen J, Serup P, Krull J, Gradwohl G et al. Opposing actions of Arx and Pax4 in endocrine pancreas development. Genes Dev 2003; 17: 2591–2603.
Venters S, Thorsteinsdottir S, Duxson M . Early development of the myotome in the mouse. Dev Dyn 1999; 216: 219–232.
Kelly A, Zacks S . The histogenesis of rat intercostal muscle. J Cell Biol 1969; 42: 135–153.
Walro J, Kucera J . Why adult mammalian intrafusal and extrafusal fibers contain different myosin heavy-chain isoforms. Trends Neurosci 1999; 22: 180–184.
Kablar B, Krastel K, Tajbakhsh S, Rudnicki M . Myf5 and MyoD activation define independent myogenic compartments during embryonic development. Dev Biol 2003; 258: 307–318.
Lassar A, Davis R, Wright W, Kadesch T, Murre C, Voronova A et al. Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47-like proteins in vivo. Cell 1991; 66: 305–315.
Molkentin J, Black B, Martin J, Olson E . Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell 1995; 83: 1125–1136.
Weintraub H, Davis R, Lockshon D, Lassar A . MyoD binds cooperatively to two sites in a target enhancer sequence: occupancy of two sites is required for activation. Proc Natl Acad Sci USA 1990; 87: 5623–5627.
Braun T, Bober E, Buschhausen-Denker G, Kohtz S, Grzeschik K, Arnold H . Differential expression of myogenic determination genes in muscle cells: possible autoactivation by the Myf gene products. EMBO J 1989; 8: 3617–3625.
Mootoosamy R, Dietrich S . Distinct regulatory cascades for head and trunk myogenesis. Development 2002; 129: 573–583.
Stromme P, Mangelsdorf M, Shaw M, Lower K, Lewis S, Bruyere H et al. Mutations in the human ortholog of Aristaless cause X-linked mental retardation and epilepsy. Nat Genet 2002; 30: 441–445.
Cserjesi P, Olson E . Myogenin induces the myocyte-specific enhancer binding factor MEF-2 independently of other muscle-specific gene products. Mol Cell Biol 1991; 11: 4854–4862.
Collombat P, Hecksher-Sorensen J, Broccoli V, Krull J, Ponte I, Mundiger T et al. The simultaneous loss of Arx and Pax4 genes promotes a somatostatin-producing cell fate specification at the expense of the alpha- and beta-cell lineages in the mouse endocrine pancreas. Development 2005; 132: 2969–2980.
McKenzie O, Ponte I, Mangelsdorf M, Finnis M, Colasante G, Shoubridge C et al. Aristaless-related homeobox gene, the gene responsible for West syndrome and related disorders, is a Groucho/transducin-like enhancer of split dependent transcriptional repressor. Neuroscience 2007; 146: 236–247.
Seufert D, Prescott N, El-Hodiri H . Xenopus aristaless-related homeobox (xARX) gene product functions as both a transcriptional activator and repressor in forebrain development. Dev Dyn 2005; 232: 313–324.
Berkes C, Bergstrom D, Penn B, Seaver K, Knoepfler P, Tapscott S . Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol Cell 2004; 14: 465–477.
Edmondson D, Cheng T, Cserjesi P, Chakraborty T, Olson E . Analysis of the myogenin promoter reveals an indirect pathway for positive autoregulation mediated by the muscle-specific enhancer factor MEF-2. Mol Cell Biol 1992; 12: 3665–3677.
Fujisawa-Sehara A, Nabeshima Y, Komiya T, Uetsuki T, Asakura A, Nabeshima Y . Differential trans-activation of muscle-specific regulatory elements including the mysosin light chain box by chicken MyoD, myogenin, and MRF4. J Biol Chem 1992; 267: 10031–10038.
Rawls A, Valdez M, Zhang W, Richardson J, Klein W, Olson E . Overlapping functions of the myogenic bHLH genes MRF4 and MyoD revealed in double mutant mice. Development 1998; 125: 2349–2358.
Cossu G, Zani B, Coletta M, Bouche M, Pacifici M, Molinaro M . In vitro differentiation of satellite cells isolated from normal and dystrophic mammalian muscles. A comparison with embryonic myogenic cells. Cell Differ 1980; 9: 357–368.
Minasi M, Riminucci M, DeAngelis L, Borello U, Berarducci B, Innocenzi A et al. The meso-angioblast: a multipotent, self-renewing cell that originates from the dorsal aorta and differentiates into most mesodermal tissues. Development 2002; 129: 2773–2783.
Colombo E, Galli R, Cossu G, Gecz J, Broccoli V . Mouse orthologue of ARX, a gene mutated in several X-linked forms of mental retardation and epilepsy, is a marker of adult neural stem cells and forebrain GABAergic neurons. Dev Dyn 2004; 231: 631–639.
Cornelison D, Olwin B, Rudnicki M, Wold B . MyoD(−/−) satellite cells in single-fiber culture are differentiation defective and MRF4 deficient. Dev Biol 2000; 224: 122–137.
Russo S, Tato F, Grossi M . Transcriptional down-regulation of myogenin expression is associated with v-ras-induced block of differentiation in unestablished quail muscle cells. Oncogene 1997; 14: 63–73.
Kelly R, Alonso S, Tajbakhsh S, Cossu G, Buckingham M . Myosin light chain 3F regulatory sequences confer regionalized cardiac and skeletal muscle expression in transgenic mice. J Cell Biol 1995; 129: 383–396.
Rudnicki M, Braun T, Hinuma S, Jaenisch R . Inactivation of MyoD in mice leads to up-regulation of the myogenic HLH gene Myf-5 and results in apparently normal muscle development. Cell 1992; 71: 383–390.
Tajbakhsh S, Rocancourt D, Buckingham M . Muscle progenitor cells failing to respond to positional cues adopt non-myogenic fates in myf-5 null mice. Nature 1996; 384: 266–270.
Messina G, Blasi C, Rocca SL, Pompili M, Calconi A, Grossi M . p27Kip1 acts downstream of N-cadherin-mediated cell adhesion to promote myogenesis beyond cell cycle regulation. Mol Biol Cell 2005; 16: 1469–1480.
Acknowledgements
The authors thank JR Beauchamp and S Hughes for helpful discussions. A particular acknowledgement also to G Lamorte, C Pardini, C Omes, V Musella, D Mascherpa, E Roncaglia for technical assistance and to M Buckingham and M Grossi for providing mutant mice and reagents. This work was supported by Muscular Dystrophy Association, Association Francoise contra les Myopathies, Duchenne Parent Project, AIRC, PRIN, the European Community (EuroStemCell and Cellsintoorgans), the Italian Ministry of University and Research and grants from the Pasteur Institute.
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by R De Maria
Rights and permissions
About this article
Cite this article
Biressi, S., Messina, G., Collombat, P. et al. The homeobox gene Arx is a novel positive regulator of embryonic myogenesis. Cell Death Differ 15, 94–104 (2008). https://doi.org/10.1038/sj.cdd.4402230
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.cdd.4402230
Keywords
This article is cited by
-
Copy number variants in patients with intellectual disability affect the regulation of ARX transcription factor gene
Human Genetics (2015)
-
Gene expression profiling of the hyperplastic growth zones of the late trout embryo myotome using laser capture microdissection and microarray analysis
BMC Genomics (2013)