Abstract
Livin, a member of the inhibitor of apoptosis protein (IAP) family, encodes a protein containing a single baculoviral IAP repeat (BIR) domain and a COOH-terminal RING finger domain. It has been reported that Livin directly interacts with caspase-3 and -7 in vitro and caspase-9 in vivo via its BIR domain and is negatively regulated by Smac/DIABLO. Nonetheless, the detailed mechanism underlying its antiapoptotic function has not yet been fully characterized. In this report, we provide, for the first time, the evidence that Livin can act as an E3 ubiquitin ligase for targeting the degradation of Smac/DIABLO. Both BIR domain and RING finger domain of Livin are required for this degradation in vitro and in vivo. We also demonstrate that Livin is an unstable protein with a half-life of less than 4 h in living cells. The RING domain of Livin promotes its auto-ubiquitination, whereas the BIR domain is likely to display degradation-inhibitory activity. Mutation in the Livin BIR domain greatly enhances its instability and nullifies its binding to Smac/DIABLO, resulting in a reduced antiapoptosis inhibition. Our findings provide a novel function of Livin: it exhibits E3 ubiquitin ligase activity to degrade the pivotal apoptotic regulator Smac/DIABLO through the ubiquitin–proteasome pathway.
Similar content being viewed by others
Introduction
In recent years, an increasing amount of attention has been given to a class of antiapoptotic proteins, the IAPs (inhibitor of apoptosis proteins), owing to their highly specialized roles in the process of apoptosis. IAPs constitute a family of highly conserved apoptotic suppressor proteins and have been found in a wide variety of species ranging from insects to human beings throughout evolution.1, 2 Members of the IAP family are characterized by 1–3 tandem baculovirus IAP repeat (BIR) motifs and some of them also possess a RING finger motif at their carboxyl-termini. IAP proteins prevent cell death by binding to and inhibiting active caspases. They are negatively regulated by IAP-binding proteins, such as mitochondrial protein Smac/DIABLO, which alleviates the IAP–caspase interaction, thus freeing the caspase for its downstream apoptotic activity.3, 4 Mature Smac/DIABLO was reported to interact with XIAP, cIAP-1, cIAP-2, Survivin or Livin to inhibit their antiapoptotic activity.5, 6, 7, 8, 9 Although RING domains have been identified in various proteins with different functions such as DNA and protein interactions,10 the most prominent role the RING domain in XIAP, cIAP-1 or cIAP-2 plays has been attributed to its E3 ubiquitin ligase activity: promoting the degradation of both targeted proteins and themselves through ubiquitination. Although XIAP and cIAP-1 have been shown to mediate their self-degradation during apoptosis of thymocytes, XIAP, cIAP-1 and cIAP-2 have also been attested as ubiquitin-protein ligase E3 for Smac/DIABLO.11, 12, 13 Additionally, whereas the cIAP-2 RING domain was shown to promote monoubiquitination of caspase-7 and caspase-3 in vitro,14 XIAP regulates proteasomal degradation of caspase-3 through polyubiquitination thereby enhancing its antiapoptotic effect on Fas-induced cell death.15 It has not yet been demonstrated, however, if this is generally true for all RING domain-containing IAP family members. Livin, also called KIAP and ML-IAP,16, 17, 18 encodes a protein with a single BIR domain and a carboxy-terminal RING domain. Recent reports have demonstrated that Livin can interact via its BIR domain with caspase-3 and -7 in vitro and caspase-9 and Smac/DIABLO in vivo.9, 16 Disruption of interaction between Livin and Smac/DIABLO abrogated the ability of Livin to inhibit apoptosis. Yang and Du19 reported that although Smac/DIABLO can efficiently promote the auto-ubiquitination of Livin, it was unable to target Livin for rapid degradation in HeLa cells. Although Livin was reported to be a poor caspase inhibitor, it can nonetheless protect cells from apoptosis induced by multiple death stimuli such as FADD, Bax, RIP, RIP3 and DR6.16 This gives rise to the possibility that Livin may exert its antiapoptotic properties by as yet uncharacterized caspase-independent mechanisms.
In this report, we provide the first evidence that Livin can act as a ubiquitin ligase E3 for targeting the degradation of Smac/DIABLO during the prevention of etoposide-induced cell death. The half-life of short-lived Livin was determined to be less than 4 h in the presence of cycloheximide (CHX), a potent eucaryotic translation inhibitor. We demonstrate that Livin-mediated polyubiquitination of Smac/DIABLO depends on the presence of both its functional RING domain and its intact BIR domain through which Smac/DIABLO interacts. Binding of Livin to Smac/DIABLO is the prerequisite for targeted ubiquitination of Smac/DIABLO. Moreover, Livin's RING domain is shown to promote its auto-ubiquitination, as RING-deleted Livin shows little, if any auto-ubiquitination. In contrast, mutations in the BIR domain of Livin do not greatly affect its self-ubiquitination in vivo, but rather significantly increase its instability. Finally, mutations in Livin's BIR domain abrogate its capability to bind to Smac/DIABLO for degradation in vivo, resulting in the reduction of its antiapoptotic inhibition. In conclusion, in addition to inhibiting caspase activity, Livin plays an alternative role in targeting the degradation of the death-promoting factor Smac/DIABLO through the ubiquitin–proteasome pathway.
Results
Livin is a short-lived protein and is degraded by the ubiquitin–proteasome pathway
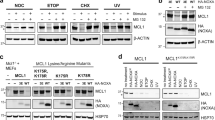
Livin, a member of the IAP family, was reported to be an unstable protein.19 To determine the half-life of this antiapoptotic factor, CHX (20 μg/ml) was added to HeLa cells and incubation was continued for indicated periods of time before cell extracts were collected and analyzed by Western blot. As shown in Figure 1a-1, endogenous Livin protein levels were dramatically decreased in the presence of CHX and approximately half of the Livin was shown to be degraded after 4 h CHX treatment. This conclusion was further confirmed by scanning densitometry and the plot is shown in Figure 1a-2. Nearly no Livin was detectable after 8 h of CHX treatment, indicating that in the absence of de novo protein synthesis, endogenous Livin degrades rapidly and its half-life (t1/2) is approximately 4 h. The faint band just above Livin marked as Livin* is expected to be its modified form, although this awaits further characterization. Next, we examined whether Livin was subjected to ubiquitin-mediated proteasomal degradation. As shown in Figure 1b-1, degradation of Livin can be blocked by proteasome inhibitor MG132 or ALLN, suggesting that the inhibition of degradation of Livin is specific for MG132 and ALLN and hence Livin is a target for proteasomal degradation. The respective densitometry data are shown in Figure 1b-2. A baseline experiment showing accumulation of Livin with proteasome inhibitor MG132 treatment in the absence of CHX was also performed. As shown in Figure 1c-1, the protein level of Livin increases gradually over the indicated period of MG132 treatment. The densitometry data are shown in Figure 1c-2. These data further support the conclusion that Livin is subjected to ubiquitin-mediated proteasome degradation.
Determining the half-life and its degradation pathway for Livin. (a-1) HeLa cells cultured in 24-well plates were treated with CHX (20 μg/ml) for 0, 2, 4, 6, 8, 10, 12 and 14 h individually before soluble extracts were collected. The relative protein levels of Livin were analyzed by Western blot with anti-Livin antibody and endogenous Actin was used as loading control. Star denotes the modified form of Livin. (b-1) Overnight-cultured HeLa cells were pretreated with DMSO (5 μl/ml), MG132 (20 μM) and ALLN (100 μM) for 1 h before the treated cells were further incubated with CHX (20 μg/ml) for the indicated periods of time. Cell lysates were then subjected to SDS-PAGE and immunoblotting with anti-Livin antibody. Endogenous Actin was used as loading control. (c-1) HeLa cells were first cultured overnight and then treated with MG132 (20 μM) for 0, 2, 4, 6, 8 and 10 h individually before soluble extracts were collected in the absence of CHX. The relative protein levels of Livin were analyzed by Western blot with anti-Livin antibody and endogenous Actin was used as loading control. (a-2, b-2, c-2) Scanning densitometry was performed for each Western blot using commercial software provided by Eaglesight (Invitrogen). Values represent means±S.D. from three independent experiments
Auto-ubiquitination of Livin in vivo
To determine the involvement of self-ubiquitination of endogenous Livin in vivo, HeLa cells were treated with or without proteasome inhibitors MG132 or ALLN for 12 h before cells were collected. As shown in Figure 2a, high-molecular-weight Livin products characteristic of polyubiquitinated products were accumulated only in cells treated with MG132 or ALLN, but not in cells left untreated, indicating that Livin is subject to polyubiquitination modification under in vivo condition (Figure 2a).
Auto-ubiquitination of Livin in vivo. (a) Overnight-cultured HeLa cells were treated with and without MG132 (20 μM) or ALLN (100 μM) for another 24 h. Cell lysates were then prepared and subjected to immunoblotting with anti-Livin antibody. Polyubiquitinated Livin is indicated as Livin-Ub(n) at the right side of the figure. (b) Schematic diagrams of pcDNA3/Flag-Livin fusion constructs for pcDNA3/Flag-Livin, pcDNA3/Flag-Livin-C124A, pcDNA3/Flag-Livin-C252A and pcDNA3/Flag-LivinΔRING. BIR domain is indicated by box in light gray and RING domain in dark gray. Numbers indicate the positions of amino-acid residues in Livin. (c) HeLa cells were transfected with pcDNA3/Flag, pcDNA3/Flag-Livin, pcDNA3/Flag-LivinΔRING and pcDNA3/Flag-Livin-C124A individually (Lanes 1–4). Twenty-four hours after transfection, cells were treated with MG132 (20 μM) for another 12 h. Cell lysates were prepared and incubated with mouse anti-Flag antibody bound to Protein A/G-Sepharose. After 6 h incubation at 4°C, the immunoprecipitates were washed five times in lysis buffer, and proteins were recovered by boiling beads in SDS sample buffer and analyzed by Western blotting using rabbit anti-Flag antibody
To further examine the effects of BIR domain and RING domain of Livin on its self-ubiquitination, four Flag-tagged Livin fusion plasmids were constructed as shown in Figure 2b. HeLa cells expressing Flag-control, Flag-Livin, Flag-Livin-C124A or Flag-LivinΔRING, respectively, were treated with MG132 for 12 h before they were collected. Cell lysates were further used for immunoprecipitation and Western analysis. As shown in Figure 2c (lanes 1–4), a high-molecular-weight smear characteristic of polyubiquitylated products was detected in cells expressing Flag-Livin and BIR mutant Flag-Livin-C124A, but not in cells expressing RING domain deletion mutant Flag-LivinΔRING, indicating that the RING domain, but not the BIR domain, of Livin is involved in its polyubiquitylation in vivo. Immunoprecipitates used for detecting polyubiquitination of Livin by anti-Flag antibody were also examined by anti-Livin antibody, and a similar result was obtained (data not shown).
We notice from Figure 2c that the level of Livin BIR mutant Flag-Livin-C124A is much lower than that of wild-type (wt)-Livin or RING deletion mutant Flag-LivinΔRING. Therefore, we examined whether the BIR domain affects the stability of Livin. HeLa cells were transfected with pcDNA3/Flag-Livin, pcDNA3/Flag-LivinΔRING, pcDNA3/Flag-Livin-C124A or pcDNA3/Flag-Livin-C252A. The reason for additionally choosing Cys252 as a mutation residue, which is underlined in the Livin RING domain CX2CX9–39CX1–3HX2–3C/HX2-C-X4–48CX2C, is the point that mutation at Cys252 (zinc-binding residues) was reported to abolish the capability of ubiquitination of Livin.19, 20 Protein stability was compared among wt-Livin, LivinΔRING, Livin-C124A and Livin-C252A with and without MG132 treatment. As shown in Figure 3, although the steady-state level of wt-Livin or Livin-C124A was considerably higher in the presence (lanes 2, 6) than in the absence of MG132 (lanes 1, 5) (densitometry data not shown), both RING deletion mutant LivinΔRING and RING point mutant Livin-C252A were expressed at comparable levels before and after MG132 treatment (lanes 3, 4, 7, 8), indicating that the RING domain, but not BIR domain, is responsible for Livin's ubiquitin-mediated proteasome degradation. We noticed that BIR mutant Livin-C124A always expressed at a lower level than wt-Livin, and moreover, MG132 treatment could not bring the level of BIR mutant back to a level similar to that of wt-Livin. Based on these observations, we explain it in more detail below: there might be two possibilities for explaining the accelerated instability of Livin BIR mutant: one, Livin BIR mutants may undergo an additional mechanism independent of proteasomal degradation (if only proteasomal degradation is involved, MG132 is expected to bring back the level of BIR mutant to the wt-Livin level, provided that BIR mutant expression level or structural stability is similar to wt-Livin); the alternative possibility is that Livin BIR mutants per se may have less expression level as a result of C124A point mutation.
Livin RING domain affects its auto-ubiquitination and protein stability. HeLa cells were cotransfected with 0.5 μg of pEGFP-C1 (transfection efficiency index) plus either pcDNA3/Flag-Livin (0.5 μg), pcDNA3/Flag-LivinΔRING (0.5 μg), pcDNA3/Flag-Livin-C124A (0.5 μg) or pcDNA3/Flag-Livin-C252A (0.5 μg). Twenty-four hours after transfection, cells in each well were split into two equal parts and one part was treated with MG132 (20 μM) and the other part was left untreated for another 12 h. Expression levels of four examined Flag/Livin fusion proteins were compared by densitometry technique in the presence and absence of MG132 (upper panel, data from densitometry scanning not shown). Beta-actin was used as loading control (lower panel)
Mutation in Livin BIR domain enhances its degradation and results in reduced antiapoptotic inhibition
To explore the role of Livin BIR domain in mediating protein stability, we constructed two additional Livin point mutants, namely Livin-W134A and Livin-H144A. Trp134 of Livin is highly conserved in IAP family and His144 of Livin corresponds to His220 of XIAP and His77 of Survivin.21, 22 Based on this analogy, amino-acid residues Cys124, Trp134 and His144 of Livin are predicted to be critical for stabilizing the overall folding of the Livin molecule. We further compared the stability of Livin BIR domain mutants with that of wt-Livin in cells expressing Livin-C124A, Livin-W134A, Livin-H144A and wt-Livin. As shown in Figure 4a-1, the steady-state level of each BIR domain mutant protein in transfected cells was much lower than that of wt-Livin in the absence of MG132 (lanes 1, 3, 5, 7, densitometry data not shown), implying that BIR mutants are less stable than wt-Livin. Nevertheless, upon treatment with MG132, their levels were much increased (lanes 2, 4, 6, 8, densitometry data not shown). However, in the presence of MG132, levels of each BIR mutant (lanes 2, 4, 6) were still lower than that of wt-Livin (lane 8, densitometry data not shown).
Livin BIR domain mutant fails to interact with Smac/DIABLO, resulting in a reduction of its antiapoptosis and an increase of its degradation. (a-1) HeLa cells were cotransfected with 0.5 μg of pEGFP-C1 (transfection efficiency index) plus either pcDNA3/Flag-Livin (0.5 μg), pcDNA3/Flag-Livin-C124A (0.5 μg), pcDNA3/Flag-Livin-W134A (0.5 μg) or pcDNA3/Flag-Livin-H144A (0.5 μg). Twenty-four hours after transfection, cells were split into two equal parts and one part was treated with MG132 (20 μM) and the other part was left untreated for another 12 h. Cells were then collected and analyzed by Western blotting using anti-Flag antibody (upper panel). Whole-cell lysates were used for detection of endogenous Actin to ensure equal sample loading per lane (lower panel). (a-2) To keep the expression level of BIR mutant and wt-Livin the same, different plasmid concentrations were used. HeLa cells were cotransfected with pEGFP/C1-RIP3 (0.5 μg) and either pcDNA3/Flag (0.5 μg), pcDNA3/Flag-Livin (0.5 μg), pcDNA3/Flag-Livin-C124A (1.5 μg), pcDNA3/Flag-Livin-W134A (1 μg) or pcDNA3/Flag-Livin-H144A (1.5 μg), respectively for 24 h and were then stained with Hoechst 33342 (5 μg/ml) for 10 min. The viability of the cells was determined by counting both the living and the dead GFP-positive cells. Five different microscopic fields containing 100–150 cells from each well were chosen at random for counting samples. Values are mean±S.D. from three independent experiments. Statistical analysis was by t-test with *P<0.05. (a-3) Cell lysates were used for detection the expression of transfected Flag-Livin and Livin mutants with anti-Flag antibody and the expression of GFP-RIP3 with anti-GFP antibody. The cleavage of pro-caspase-9 was detected by anti-caspase-9 antibody and the cleaved products were indicated as 35 and 37 kDa, respectively. The cleavage of PARP was detected by anti-PARP antibody and the cleaved product was indicated as 85 kDa. Actin was used as loading control. (b-1) HeLa cells were cultured overnight and then treated with and without Taxol (200 nM) or etoposide (100 μg/ml) for 24 h. Cytosolic fraction prepared from each sample was immunoprecipitated by anti-Smac antibody and the immunoprecipitates were analyzed by Western blotting using anti-Livin antibody. Cell lysates were also used for detection of endogenous Livin by anti-Livin antibody. (b-2) HeLa cells were transfected with pcDNA3/Flag (0.5 μg), pcDNA3/Flag-Livin (0.5 μg), pcDNA3/Flag-Livin-C124A (0.5 μg), pcDNA3/Flag-Livin-W134A (0.5 μg), pcDNA3/Flag-Livin-H144A (0.5 μg) or pcDNA3/Flag-LivinΔRING (0.5 μg) for 24 h before each transfectant was split into two equal parts. One part was treated with Taxol (200 nM) and the other part was left untreated. Twenty-four hours later, cytosolic fractions prepared from each transfectant were then incubated with anti-Smac antibody-loaded Protein A/G-Sepharose for 6 h at 4°C, and the immunoprecipitates were analyzed by Western blotting using anti-Flag antibody (upper panel). Cell lysates were also used for detection of the expression of respective transfected Flag-Livin fusions by anti-Flag antibody (middle panel). The presence of Smac/DIABLO in each immunoprecipitate precipitated by anti-Smac antibody was confirmed by Western analysis (bottom panel)
If the notion that mutations in the BIR domain enhance Livin's degradation was true, we would expect that cells expressing Livin BIR mutant would be more prone to apoptosis than cells expressing wt-Livin. Kasof and Gomes16 reported that Livin was able to prevent HeLa cells from apoptosis induced by RIP3; we therefore compared the antiapoptotic ability of wt-Livin and Livin BIR mutants in preventing RIP3-induced apoptosis. As shown in Figures 3, 4a-1, the BIR mutants expression level is lower than that of the wt-Livin. In order to have a better comparison of their apoptotic inhibition, we adjusted the amount of three BIR mutant plasmids to the same expression level as wt-Livin (Figure 4a-3, second panel). HeLa cells were cotransfected with pEGFP/C1-RIP3 plus either pcDNA3/Flag-Livin (0.5 μg), pcDNA3/Flag-Livin-C124A (1.5 μg), -W134A (1 μg) or -H144A (1.5 μg) individually. Transfected cells were stained with Hoechst 33342 and the viability of the cells was scored and compared (Figure 4a-2). As expected, wt-Livin (31.53%) was shown to significantly reduce RIP3-induced apoptosis compared with vector control (71.72%). In contrast, three BIR domain mutants, namely Livin-C124 (57.41%), -W134 (49.42%) and -H144 (48.48%), showed diminished antiapoptotic function (P<0.05, t-test), indicating that an intact BIR domain is essential for Livin's protective role. The cleavage of pro-caspase-9 and PARP representing exertion of apoptosis was detected in RIP3-induced apoptosis (Figure 4a-3), and the cleavage degree of procaspase-9 and PARP is in accordance with that of apoptosis shown in Figure 4a-2.
To explicate whether the reduced antiapoptotic function of Livin BIR mutants could be attributed to the loss of their ability to bind Smac/DIABLO, we first verified that Livin interacts with Smac/DIABLO in living cells. Immunoblotting results revealed that anti-Smac antibody could only co-immunoprecipitate (Co-IP) with endogenous Livin in the cells treated with drugs, but not in those cells left untreated (Figure 4b–1). We then examined whether Smac/DIABLO can interact with Livin mutants. As shown in Figure 4b-2, in the presence of Taxol treatment, anti-Smac antibody could only Co-IP wt-Livin and LivinΔRING, but not any of the Livin BIR mutants (Figure 4b–2, the left-hand side of the top panel). As a control, neither wt-Livin nor mutant Livin proteins could be co-immunoprecipitated with Smac/DIABLO by anti-Smac antibody without Taxol treatment (Figure 4b-2, the right-hand side of the top panel), suggesting that the association of Smac/DIABLO and Livin is dependent on Taxol or etoposide treatment. In conclusion, the Livin BIR domain is crucial for its binding to Smac/DIABLO and mutation in the BIR domain nullifies this interaction, and hence reduces Livin's antiapoptotic function.
Smac/DIABLO is a ubiquitination substrate for Livin
We have demonstrated that Livin physically interacts with Smac/DIABLO through its BIR domain. To further investigate whether Livin is able to target Smac/DIABLO for degradation and hence decreases the level of Smac/DIABLO in favor of cell survival, a Western analysis was performed. As shown in Figure 5a, in the absence of MG132, the protein level of cytosolic Smac/DIABLO was significantly decreased in cells expressing Flag-Livin (lane 3) when compared with cells expressing Flag control (lane 1), Flag-LivinΔRING (lane 5) or Flag-Livin-C124A (lane 7). In the presence of MG132, however, levels of Smac/DIABLO become comparable in all four transfected cells, implying that (i) Smac/DIABLO appeared to be targeted for degradation by Livin, but not by Livin-RING domain or -BIR domain mutant, and (ii) this Livin-targeted degradation is proteasome-mediated.
Smac/DIABLO is a ubiquitination substrate for Livin in vivo and in vitro. (a) HeLa cells were transiently cotransfected with 0.5 μg of pEGFP-C1 (transfection efficiency index) plus either pcDNA3/Flag (0.5 μg), pcDNA3/Flag-Livin (0.5 μg), pcDNA3/ Flag-LivinΔRING (0.5 μg) or pcDNA3/Flag-Livin-C124A (0.5 μg). Twenty-four hours after transfection, cells in each well were split into two equal parts and cultured for a short period of time. All the samples that were split were first treated with etoposide (100 μg/ml) for 24 h, followed by treatment with or without MG132 (20 μM) for an additional 12 h. Cytosolic fractions were prepared for detection of the steady-state level of cellular Smac/DIABLO by Western blotting using anti-Smac antibody (top panel). The successful expression of transfected Flag-Livin fusions was verified by anti-Flag antibody (middle panel). Detection of endogenous beta-actin was used as loading control (bottom panel). (b) Cells were stained with Hoechst 33342 and both live and dying/dead cells were scored and the data were plotted as percent apoptosis. Each bar represents the mean±S.D. from three independent experiments. Statistical analysis was by t-test with *P<0.05. (c) HeLa cells were transiently transfected with pcDNA3/Flag, pcDNA3/Flag-LivinΔRING, pcDNA3/Flag-Livin-C124A or pcDNA3/Flag-Livin (lanes 1–4). Twenty-four hours after transfection, cell were treated with etoposide (20 μM) for another 12 h followed by treating with or without 10 μM MG132 for an additional 12 h. Cytosolic fractions were immnoprecipitated by anti-Smac antibody (mouse monoclonal antibody from Cell Signaling Technology) and the immunoprecipitates were further analyzed by Western blotting using anti-ubiquitin antibody (rabbit polyclonal antibody from Calbiochem) and donkey anti-rabbit IgG (from Amersham) (left upper panel) and anti-Smac antibodies (right panel). The same cytosolic fractions were used for detection of expression of various transfected Flag-Livin and Flag-Livin mutants (left lower panel). (d) Assays were performed in the presence of all the other assay components, but in the absence of purified recombinant His-Smac, E1, E2, GST-Livin for 120 min (lanes 1–4). The effect of incubating for 2 h at 30°C in the presence of purified GST, GST-Livin, GST-Livin-C124A or GST-LivinΔRING individually is shown in lanes 5–8. Assay reaction buffer utilized contained 50 mM Tris-HCl (pH 7.6), 100 nM rabbit E1 and 400 nM E2 UbcH5b (in E2 titration experiment, 200 nM and 100 nM E2 were also used, data not shown), 20 μM ubiquitin and 2 mM Mg-ATP. When the assay was completed, SDS was added to stop the reaction followed by Western blotting using anti-Smac antibody. High-molecular-weight polyubiquitinated Smac/DIABLO is marked as Smac-Ub(n) on the right side of the figure
A reduced level of Smac/DIABLO is expected to decrease its apoptotic potential. Apoptosis induced by etoposide was measured in four above-mentioned transfected cells and the corresponding results are plotted in Figure 5b. In the absence of MG132, apoptosis of transfectants induced by etoposide ordered increasing is as follows: cells expressing Flag-Livin (37.17%), Flag-LivinΔRING (49.43%) and Flag-Livin-C124A (56.27%) (P<0.05, t-test). This result is in agreement with the hypothesis that reduced degradation of Smac/DIABLO by Livin mutant could result in increased apoptosis.
Next, we examined whether Livin truly ubiquitinates Smac/DIABLO for degradation in vivo. An immunoprecipitation experiment was performed. As shown in Figure 5c, Smac/DIABLO became heavily polyubiquitinated in those cells expressing wt-Livin, whereas cells expressing either Livin BIR mutant (Livin-C124A) or RING mutant (LivinΔRING) showed little, if any background ubiquitination of Smac/DIABLO. Immunoprecipitates used for detecting polyubiquitination by anti-ubiquitin antibody were also examined for the presence of Smac/DIABLO by anti-Smac antibody (Figure 5c, middle panel). In conclusion, Livin-mediated ubiquitination of Smac/DIABLO in vivo depends on the presence of both an intact Livin RING finger domain and its functional BIR domain.
Lastly, to provide more convincing evidence that Livin by itself can ubiquitinate Smac/DIABLO, an in vitro experiment was performed. As shown in Figure 5d, Smac/DIABLO was found to be ubiquitinated by GST-Livin (lane 8), but not GST-LivinRING or GST-Livin-C124A (lanes 6, 7), indicating that BIR domain and RING domain are essential for Livin's E3 ligase activity under in vitro conditions. However, if LivinRING is in excess, some degree of polyubiquination of Smac/DIABLO by LivinRING can be observed (data not shown). It is well known that excessive E2 can substitute for the requirement of an E3 in an in vitro assay; we therefore titrated down the concentration of E2 (UbcH5b) from 400 nM (concentration used in in vitro assay) down to 200 nM and 100 nM and found that ubiquitination of Smac/DIABLO is, at least in vitro, independent of E2 concentration (data not shown). We concluded that Smac/DIABLO ubiquitylation in vitro by Livin requires the RING domain. This conclusion is in accordance with the results in vivo.
Discussion
Among many mechanisms that regulate the activity of IAP, post-translational modifications, particularly ubiquitination, have received increasing attention. To investigate whether IAP protein Livin, like other RING finger proteins, can regulate itself through auto-ubiquitination,11, 23, 24 we compared the endogenous Livin in the presence or absence of proteasome inhibitor MG132 or ALLN. We found that Livin is regulated by self-ubiquitination, and as a result, Livin undergoes proteasome-mediated degradation. A previous report demonstrated that although processed mature Smac/DIABLO is able to promote auto-ubiquitination of Livin, it has little, if any, effect on its degradation.19 It is therefore reasonable to assume that Livin RING domain could be the candidate determinant of its self-ubiquitination. Data from our experiment proved that the RING domain of Livin plays a crucial role in its auto-ubiquitination, as Livin RING deletion mutant demolishes all its auto-ubiquitination activity (Figure 2c). However, we cannot completely exclude the possibility that Livin auto-ubiquitination is partially enhanced by processed mature Smac/DIABLO owing to longer MG132 or ALLN treatment.
We demonstrated that Livin BIR mutant Livin-C124A showed much less stability compared with wt-Livin (Figure 2c), implying that the BIR domain per se may normally possess suppressing function for protein degradation or, alternatively, some yet uncharacterized degradation-inhibitory factor regulates its stability through the BIR domain, and mutation in the BIR domain thereby occurs, resulting in its decreased stability. However, we found that the BIR mutant exhibits less auto-ubiquitination than wt-Livin, which may be explained by the possibility that Livin BIR mutant is degraded by an as yet uncharacterized mechanism that is independent of the auto-ubiquitination process. However, the possibility that BIR mutant has a lower expression level than wt-Livin cannot be excluded at the present time. Nevertheless, we have characterized some critical amino-acid residues within the BIR domain, which greatly sensitize Livin to ubiquitin–proteasome degradation. Vucic et al.18 reported that Livin-C124A could no longer block Fas- and TNFR1-induced apoptosis, as C124 together with H144 is predicted to chelate zinc ions to stabilize the overall structure of Livin.18 In the present study, we were able to demonstrate that Livin BIR domain point mutant Livin-C124A, Livin-W134A or Livin-H144A degrades more rapidly than wt-Livin. Accordingly, all three Livin BIR domain mutants showed less effectiveness in inhibiting RIP3-induced apoptosis, supporting the conclusion that Livin's antiapoptotic activity is dependent on its intact BIR domain.
Thus far, no obvious role has been attributed to W67 in Survivin or W210 in XIAP for their antiapoptotic activity. We firstly showed that W134 of Livin (which corresponds to W67 in Survivin and W210 in XIAP) is functionally important, and that Livin BIR domain point mutant Livin-W134A not only decreases its ability to protect cells from RIP3-induced apoptosis, but also blocks interaction with mitochondria death inducer Smac/DIABLO. This suggests that W134, like C124 and H144, is a critical residue for Livin to bind Smac/DIABLO (Figure 4b-2). From these discussions, we conclude that integrity of BIR domain stabilizes Livin and enables it to exert its antiapoptotic activity.
Recent studies have shown that many RING finger-containing IAPs possess E3 activity, which appends additional biological activities to IAPs. In Drosophila melanogaster, the DIAP1 RING finger was demonstrated to mediate ubiquitination of Dronc and is indispensable for regulating apoptosis.25 In addition, XIAP and DIAP1 were reported to serve as ubiquitin ligases for Reaper and this regulation has a significant impact on the ability of Reaper to initiate apoptosis.26 Apollon was reported to function as an E2-Ubc to ubiquitinate both Smac/DIABLO and caspase-9 and thus displays an essential cytoprotection function in preventing Smac/DIABLO-induced apoptosis.27 The interacting partners of Livin include caspase-3, -7 and -9 and proteins containing an IAP-interacting motif, which currently comprise the mitochondrial protein Smac/DIABLO and Omi/HtrA2. Smac/DIABLO is a negative regulator of Livin and is therefore a proapoptotic protein. Wilkinson et al.28 reported that cytoprotective IAPs could inhibit apoptosis through the neutralization of IAP antagonists, such as Smac/DIABLO, rather than by directly inhibiting caspases. Regulation of apoptosis by Livin is believed to sequester Smac/DIABLO and prevents it from antagonizing XIAP-mediated inhibition of caspases.29 Whether Livin directs the ubiquitination of interacting proteins, such as Smac/DIABLO through its RING domain, has not yet been characterized.
We demonstrate that ectopic expression of wt-Livin, but not of Livin RING domain or Livin BIR domain mutant, promotes Smac/DIABLO ubiquitination. Wt-Livin protects cells from etoposide-induced cell death more effectively than RING deletion mutant or BIR domain point mutant does. Ubiquitination of Smac/DIABLO by Livin requires both functional BIR and RING domain in vivo and in vitro. Livin binds to Smac/DIABLO through its BIR domain and their interaction is the prerequisite for targeting the degradation of Smac/DIABLO. BIR domain point mutant Livin-C124A, which was unable to interact with Smac/DIABLO, had abrogated its ability for ubiquitinating Smac/DIABLO. Equally important, Livin RING finger domain is also indispensable for ubiquitinating Smac/DIABLO in vivo (Figure 5c). However, it should be noted that the smeared ubiquitin-modified Smac/DIABLO bands in the anti-ubiquitin or anti-Smac blots were still detected in the presence of Livin-C124A or LivinΔRING (Figure 5c, lanes 2, 3). This is not unexpected as Smac/DIABLO is also subject to polyubiquitination by other IAP factors, such as Apollon/Bruce, cIAP-1 and cIAP-2.14, 27
The struggle between cell survival and death is ubiquitous and this is maintained in balance under various intricate regulations including the mechanism of proteasome ubiquitination. On the one hand, by catalyzing its own ubiquitination, Livin lowers the threshold for inducing apoptosis and makes cells more perceptible for cell death. On the other hand, Livin targets death inducer Smac/DIABLO for degradation through the proteasome ubiquitination pathway, promoting cell survival. Understanding the detailed mechanisms underlying the dual functions of Livin in regulating cellular apoptosis and survival requires more extensive future research.
Materials and Methods
Oligonucleotides
The sequences of the oligonucleotides used in this study are listed as follows (all primers are read from 5′ to 3′):
-
P1: CGGAATTCCATGGGACCTAAAGACAGTGCCAAG;
-
P2: CGGTCGACCTAGGACAGGAAGGTGCGCACGCG;
-
P3: CAGGACAAGGTGAGGGCCTTCTTCTGCTATGGGGGC;
-
P4: AGCAGAAGAAGGCCCTCACCTTGTCCTGATG;
-
P5: GCAGAGCGCGAAGCGCGGGGACGACC;
-
P6: CCCCGCGCTTCGCGCTCTGCAGGCCCCCATA;
-
P7: GGACGGAGGCTGCCAAGTGGTTCCCCAG;
-
P8: TGGCAGCCTCCGTCCAGGGGTCG;
-
P9: GCAGGAGGAGAGGACGGCCAAGGTGTGCCTGGACCGC;
-
P10: CCAGGCACACCTTGGCCGTCCTCTCCTCCTGCAGCCGCCG;
-
P11: CGGTCGACCTACACATCCCTGGCTCCTGGGGGCTC.
Reagents and antibodies
The following antibodies were used in this study: polyclonal antibodies: anti-Actin antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA), pAb-ubiquitin (Calbiochem, La Jolla, CA, USA), pAb-anti-Flag (Sigma, St. Louis, MO, USA), pAb-anti-PARP (Upstate, Lake Placid, NY, USA). Monoclonal antibodies: mAb-Flag (Sigma), mAb-GFP (MBL, Nagoya, Japan), mAb-Smac/DIABLO (Cell Signaling Technology), mAb-caspase-9 (Immunotech, France) and mAb-Livin (IMGENEX, San Diego, CA, USA). Proteasome inhibitor MG132 (N-CBZ-leu-leu-leucinal) was purchased from Calbiochem (La Jolla, CA, USA) and ALLN (N-acetyl-leu-leu-nor leucinal, or calpain inhibitor I) was purchased from Sigma. CHX, Hoechst 33342 and etoposide were purchased from Sigma. Taxol used in our experiments was labeled as GCP (Good Clinical Practice) quality standard. Restriction enzymes were purchased from New England Biolabs (Beverly, MA, USA). Medium compounds were obtained from Oxid (Basingstroke Hampshire, UK). Rabbit ubiquitin-activating enzyme E1 and recombinant His-tagged ubiquitin were purchased from Calbiochem.
Cell culture and transfection
HeLa cells were maintained in DMEM containing 10% heat-inactivated fetal bovine serum (FBS), 1 × nonessential amino acids, 1 × MEM sodium pyruvate, 100 μg/ml penicillin and 100 μg/ml streptomycin (Invitrogen, Carlsbad, CA, USA). Cultured cells were incubated in a humidified atmosphere containing 5% CO2 at 37°C. Transfection of cells with various mammalian expression vectors by Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) was according to the methods provided by manufacturer's specification.
PCR-mediated mutagenesis
PCR-mediated mutagenesis method described previously8 was used to generate Livin point mutants including Livin-C124A, -W134A, -H144A, and -C252A. The primer pairs P1/P4 and P3/P2 were used for generating Livin-C124A; P1/P6 and P5/P2 for -W134A; P1/P8 and P7/P2 for -H144A; and P1/P10 and P9/P2 for -C252A. Mutations created were further verified by DNA sequencing.
Plasmid construction
Full-length Livin gene fragment was amplified from HeLa cDNA library by PCR using the primer pair P1/P2. The amplified fragment was subcloned into EcoRI/SalI sites of pEGFP-C1 (for the preparation of GFP/Livin) or EcoRI/XhoI sites of pCDNA3/Flag (for the preparation of Flag/Livin). A cDNA fragment coding for LivinΔRING (residues1–239) generated by PCR reaction using primers P1/P11 was digested with restriction enzymes EcoRI/SalI or EcoRI/XhoI to subclone into pEGFP-C1 and pcDNA3/Flag, respectively. PET22b-Smac (Δ55) was constructed by the method described previously.8
CHX inhibition
Protein stability was measured by using CHX as described previously.30 Briefly, HeLa cells were cultured in 24-well plates overnight at 37°C before proteasome inhibitors MG132 or ALLN were added. After incubation with MG132 or ALLN for 1 h, cells were further treated with 20 μg/ml CHX for indicated periods of time and were then harvested and analyzed by Western blotting.
Cell death assay
The ability of Livin and Livin mutants to affect cell viability was assayed by transfecting HeLa cells in 24-well plates (2 × 104 cells/well) with various Livin constructs. In order to compare the protein level and to count the dead cells more accurately, same concentration of plasmids under the same conditions used for transfection was strictly employed. Twenty-four hours after transfection, cell incubation with the drug was continued for the desired period of time and the viability of the cells was measured by counting round GFP-positive cells characterized with aberrant nuclei stained by Hoechst 33342. Data are expressed as percentages of control and are means of three independent experiments.
Western blot analysis and immunoprecipitation
Western blot analysis was performed according to the procedures described by Song et al.8 For immunoprecipitation, cells were first lysed in a Triton X-100-based lysis buffer (1% Triton X-100, 10% glycerol, 150 mM NaCl, 20 mM Tris-HCl, pH 7.5, 2 mM EDTA, protease inhibitor cocktail) for 1 h, and the nuclear and cellular debris was cleared by centrifugation. Cytosolic lysate was then incubated with first monoclonal antibody-bound Protein A/G-Sepharose. After 1 h incubation at 4°C, the immunoprecipitates were washed five times in lysis buffer, and proteins were recovered by boiling beads in SDS sample buffer and analyzed by Western blot.
In vitro ubiquitination assay
GST and GST-fusion proteins were expressed in Escherichia coli strains DH5-alpha or BL21/DE3. The GST-fusion proteins were purified through the Glutathione Sepharose 4B beads (Amersham Pharmacia Biotech, Uppsala, Sweden). Assay reaction buffer utilized for in vitro ubiquitination of Smac/DIABLO by Livin and Livin mutants contained 50 mM Tris-HCl (pH 7.6), 100 nM rabbit E1 and 400 nM E2 UbcH5b, 20 μM ubiquitin and 2 mM Mg-ATP. When the assay was completed, SDS was added to stop the reaction, followed by Western blotting using anti-Smac antibody.
Abbreviations
- IAP:
-
inhibitor of apoptosis protein
- BIR:
-
baculovirus IAP repeat
- GST:
-
glutathione S-transferase
- GFP:
-
green fluorescent protein
- EGFP:
-
enhanced green fluorescent protein
- CHX:
-
cycloheximide
- PARP:
-
poly(ADP-ribose) polymerase
- FADD:
-
Fas-associated protein with death domain
- RIP:
-
receptor-interacting protein
- RIP3:
-
receptor-interacting protein 3
- DR6:
-
death receptor 6
- XIAP:
-
X-linked IAP
- TNFR1:
-
tumor necrosis factor receptor 1
- Smac/DIABLO:
-
second mitochondria-derived activator of caspase/direct IAP-binding protein with low PI
References
Miller LK (1999) An exegesis of IAPs: salvation and surprises from BIR motifs. Trends Cell Biol. 9: 323–328.
Deveraux QL and Reed JC (1999) IAP family proteins – suppressors of apoptosis. Genes Dev. 13: 239–252.
Deveraux QL, Takahashi R, Salvesen GS and Reed JC (1997) X-linked IAP is a direct inhibitor of cell-death proteases. Nature 388: 300–304.
Roy N, Deveraux QL, Takahashi R, Salvesen GS and Reed JC (1997) The c-IAP-1 and c-IAP-2 proteins are direct inhibitors of specific caspases. EMBO J. 16: 6914–6925.
Du C, Fang M, Li Y, Li L and Wang X (2000) Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell 102: 33–42.
Verhagen AM, Ekert PG, Pakusch M, Silke J, Connolly LM, Reid GE, Moritz RL, Simpson RJ and Vaux DL (2000) Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. Cell 102: 43–53.
Ekert PG, Silke J, Hawkins CJ, Verhagen AM and Vaux DL (2001) DIABLO promotes apoptosis by removing MIHA/XIAP from processed caspase 9. J. Cell Biol. 152: 483–490.
Song ZY, Yao XB and Wu M (2003) Direct interaction between survivin and Smac/DIABLO is essential for the anti-apoptotic activity of survivin during taxol-induced apopotosis. J. Biol. Chem. 278: 23130–23140.
Vucic D, Deshayes K, Ackerly H, Pisabarro MT, Kadkhodayan S, Fairbrother WJ and Dixit VM (2002) SMAC negatively regulate the anti-apoptotic activity of melanoma inhibitor of apoptosis(ML-IAP). J. Biol. Chem. 277: 12275–12279.
Borden KL and Freemont PS (1996) The RING finger domain: a recent example of a sequence–structure family. Curr. Opin. Struct. Biol. 6: 395–401.
Yang Y, Fang S, Jensen JP, Weissman AM and Ashwell JD (2000) Ubiquitin protein ligase activity of IAPs and their degradation in proteasomes in response to apoptotic stimuli. Science 288: 874–877.
MacFarlane M, Merrison W, Bratton SB and Cohen GM (2002) Proteasome-mediated degradation of Smac during apoptosis: XIAP promotes Smac ubiquitination in vitro. J. Biol. Chem. 277: 36611–36616.
Hu S and Yang X (2003) Cellular inhibitor of apoptosis 1 and 2 are ubiquitin ligases for the apoptosis inducer Smac/DIABLO. J. Biol. Chem. 278: 10055–10060.
Huang H, Joazeiro CA, Bonfoco E, Kamada S, Leverson JD and Hunter T (2000) The inhibitor of apoptosis, cIAP2, functions as a ubiquitin-protein ligase and promotes in vitro monoubiquitination of caspases 3 and 7. J. Biol. Chem. 275: 26661–26664.
Suzuki Y, Nakabayashi Y and Takahashi R (2001) Ubiquitin protein ligase activity of X-linked inhibitor of apoptosis protein promotes proteasomal degradation of caspase-3 and enhances its anti-apoptotic effect in Fas-induced cell death. Proc. Natl Acad. Sci. USA 98: 8662–8667.
Kasof GM and Gomes BC (2001) Livin, a novel inhibitor of apoptosis protein family member. J. Biol. Chem. 276: 3238–3246.
Lin JH, Deng G, Huang Q and Morser J (2000) KIAP, a novel member of the inhibitor of apoptosis protein family. Biochem. Biophys. Res. Commun. 279: 820–831.
Vucic D, Stennicke HR, Pisabarro MT, Salvesen GS and Dixit VM (2000) ML-IAP, a novel inhibitor of apoptosis that is preferentially expressed in human melanomas. Curr. Biol. 10: 1359–1366.
Yang QH and Du C (2004) Smac/DIABLO selectively reduces the levels of c-IAP1 and c-IAP2 but not that of XIAP and Livin in Hela cells. J. Biol. Chem. 279: 16963–16970.
Creagh EM, Murphy BM, Duriez PJ, Duckett CS and Martin SJ (2004) Smac/Diablo antagonizes ubiquitin ligase activity of inhibitor of apoptosis proteins. J. Biol. Chem. 279: 26906–26914.
Sun C, Cai M, Gunasekera AH, Meadows RP, Wang H, Chen J, Zhang H, Wu W, Xu N, Ng SC and Fesik SW (1999) NMR structure and mutagenesis of the inhibitor-of-apoptosis protein XIAP. Nature 401: 818–822.
Li F, Ambrosini G, Chu EY, Plescia J, Tognin S, Marchisio PC and Altieri DC (1999) Control of apoptosis and mitotic spindle checkpoint by survivin. Nature 396: 580–584.
Joazeiro CA, Wing SS, Huang H, Leverson JD, Hunter T and Liu YC (1999) The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science 286: 309–312.
Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S and Weissman AM (1999) RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. USA 96: 11364–11369.
Wilson R, Goyal L, Ditzel M, Zachariou A, Baker DA, Agapite J, Steller H and Meier P (2002) The DIAP1 RING finger mediates ubiquitination of Dronc and is indispensable for regulating apoptosis. Nat. Cell Biol. 4: 445–450.
Olson MR, Holley CL, Yoo SJ, Huh JR, Hay BA and Kornbluth S (2003) Reaper is regulated by IAP-mediated ubiquitination. J. Biol. Chem. 278: 4028–4034.
Hao Y, Sekine K, Kawabata A, Nakamura H, Ishioka T, Ohata H, Katayama R, Hashimoto C, Zhang X, Noda T, Tsuruo T and Naito M (2004) Apollon ubiquitinates SMAC and caspase-9, and has an essential cytoprotection function. Nat. Cell Biol. 6: 849–860.
Wilkinson JC, Wilkinson AS, Scott FL, Csomos RA, Salvesen GS and Duckett CS (2004) Neutralization of Smac/Diablo by inhibitors of apoptosis (IAPs): a caspase-independent mechanism for apoptotic inhibition. J. Biol. Chem. 279: 51082–51090.
Vucic D, Franklin MC, Wallweber HJ, Das K, Eckelman BP, Shin H, Elliott L O, Kadkhodayan S, Deshayes K, Salvesen GS and Fairbrother WJ (2005) Engineering ML-IAP to produce an extraordinarily potent caspase 9 inhibitor: implications for Smac-dependent anti-apoptotic activity of ML-IAP. Biochem J. 385 (Part 1): 11–20.
Patrick GN, Zhou P, Kwon YT, Howley PM and Tsai LH (1998) P35, the neuronal-specific activator of cyclin-dependent kinase5 (cdk5) is degraded by the ubiquitin–proteasome pathway. J. Biol. Chem. 273: 24057–24064.
Acknowledgements
We are grateful to Dr. Dirk Bohmann (University of Rochester Medical Center) for generously providing the ubiquitin plasmid used in this study and Dr. P Cheung (Nanyang Technological University) for providing E2 UbcH5b. This research was supported by a 973 grant (2002CB713702) from the Ministry of Science and Technology of China, by grants from the National Natural Science Foundation of China (30530200, 30370308, 90208027 and 30121001), by an ARC grant (to MW) from Ministry of Education, Singapore and by a grant (SBS/SUG/22/04) to KH from Nanyang Technological University, School of Biological Sciences, Singapore.
Author information
Authors and Affiliations
Author notes
L Ma, Y Huang: These authors contributed equally to this work.
Corresponding author
Additional information
Edited by GM Cohen
Rights and permissions
About this article
Cite this article
Ma, L., Huang, Y., Song, Z. et al. Livin promotes Smac/DIABLO degradation by ubiquitin–proteasome pathway. Cell Death Differ 13, 2079–2088 (2006). https://doi.org/10.1038/sj.cdd.4401959
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.cdd.4401959
Keywords
This article is cited by
-
Diverse maturity-dependent and complementary anti-apoptotic brakes safeguard human iPSC-derived neurons from cell death
Cell Death & Disease (2022)
-
Dysregulated Expression of Apoptosis-Associated Genes and MicroRNAs and Their Involvement in Gastric Carcinogenesis
Journal of Gastrointestinal Cancer (2021)
-
PROteolysis TArgeting Chimeras (PROTACs) as emerging anticancer therapeutics
Oncogene (2020)
-
CYLD downregulates Livin and synergistically improves gemcitabine chemosensitivity and decreases migratory/invasive potential in bladder cancer: the effect is autophagy-associated
Tumor Biology (2016)
-
Expression and prognostic significance of livin/BIRC7 in childhood acute lymphoblastic leukemia
Medical Oncology (2014)