Abstract
β-Amyloid precursor protein binding protein 1 (APP-BP1) was previously identified based on its binding to the carboxyl terminal of β-amyloid precursor protein. In this report, we have discovered that a mutation of dAPP-BP1 (Drosophila ortholog of APP-BP1) hinders tissue development, causes apoptosis in imaginal disc cells, and blocks the NEDD8 conjugation pathway. We show that dAPP-BP1 specifically binds the intracellular domain of APP-like protein (APPL). The dAPP-BP1 mutation partially suppresses the abnormal macrochaete phenotype of Appld, while overexpression of dAPP-BP1 causes abnormal macrochaetes. When APPL is overexpressed, the normal bristle pattern in the fly thorax is disturbed and apoptosis is induced in wing imaginal discs. APPL overexpression phenotypes are enhanced by reducing the level of dAPP-BP1. APPL overexpression is shown to inhibit the NEDD8 conjugation pathway. APPL-induced apoptosis is rescued by overexpression of dAPP-BP1. Our data suggest that APPL and dAPP-BP1 interact antagonistically during Drosophila development.
Similar content being viewed by others
Main
β-Amyloid precursor protein binding protein 1 (APP-BP1) was originally identified as a protein that interacts with the intracellular carboxyl terminal of APP (β-amyloid precursor protein), thought to play a critical role in Alzheimer's disease (AD).1 APP-BP1 is homologous to the amino (N) terminus of the ubiquitin-activating enzyme E1, but lacks the conserved cysteine required for ubiquitin activation. APP-BP1 forms a heterodimer with hUba3. The hUba3 subunit is homologous to the carboxyl-terminus of E1 and apparently contributes the conserved cysteine to the dimer. The APP-BP1/hUba3 heterodimer provides E1-like functions in NEDD8 (ubiquitin-like protein) activation. Activated NEDD8 forms a thiol ester bond with the conjugating enzyme hUbc12, analogous to E2 in the ubiquitination pathway. Target proteins are covalently bound at a lysine residue.2 In mammalian cell lines, APP-BP1 drives the S- to M-phase transition in dividing cells. When overexpressed, it can cause apoptosis in primary neurons. Apoptosis can be blocked by inhibition of the NEDD8 conjugation pathway.3 In AD patients, brain APP-BP1 protein levels in lipid rafts are increased.4
Well-characterized substrates described for the NEDD8 conjugation pathway (neddylation) mainly include only members of the cullin family of proteins. Every cullin family member is known to be modified by NEDD8,5 except for APC2. Recent studies have indicated that cytoskeletal proteins, mdm2, and p53 may be additional targets for this pathway:6 in the case of p53, neddylation inhibits transcriptional activity.6 Cullins are common subunits of Roc1/Rbx1/Hrt1–Skp1–Cullin/Cdc53-F box (SCF) complexes, which regulate the stability of various proteins involved in cell cycle control and signal transduction.7, 8, 9, 10 Neddylation of cullins regulates the E3 ubiquitin ligase activity of Roc1/Rbx1/Hrt1-Skp1-Cullin/Cdc53-F box (SCF) complexes. Recent studies have shown that NEDD8 modification facilitates the recruitment of the E2 ubiquitin-conjugating enzyme to the SCF complex and mediates the dissociation of p120CAND1 an inhibitor of SCF ligase activity.11, 12 In Drosophila NEDD8 mutants, there is an accumulation of protein levels for the cell cycle regulator, Cyclin E (CycE), and for the signal transduction effectors, Cubitus interruptus (Ci), and Armadillo (Arm).13 Recently, NEDD8 has been shown to be incorporated into a variety of ubiquitinated inclusions, including the neurofibrillary tangles present in AD.14
APP is the precursor of the amyloid β (Aβ) peptide, the main component of senile plaques in AD. APP is synthesized as a transmembrane glycoprotein composed of extracellular and cytoplasmic domains.15, 16 APP undergoes proteolytic cleavage with release of an ectodomain into the extracellular space.17, 18, 19 The APP homolog in Drosophila is called APP-like protein (APPL),15 a pan-neural protein belonging to the conserved APP family. Homology among APP members is paralleled by their ability to substitute for each other functionally: transgenes encoding either Drosophila Appl or human neural App will rescue the behavioral defects of Appl null flies.20 Different forms of Drosophila APPL protein are enriched in different areas of the brain.21 Deletion of the Appl gene does not cause obvious defects in adult brain morphology.20 However, loss- and gain-of-function studies have revealed a role for APPL in synapse differentiation and axonal transport.22, 23, 24 The mutant loechrig presents with a neurodegenerative phenotype that is strongly enhanced by Appld mutants.25 Overexpression of wild-type and mutant APPL proteins has also been shown to cause abnormal neurite outgrowth.26 The cytoplasmic C-terminus of APP plays critical roles in cellular trafficking and delivery of proteases. Adaptor proteins with phosphotyrosine-binding (PTB) domains, including those in the X11, Fe65, and c-Jun N-terminal kinase (JNK)-interacting protein (JIP) families, bind specifically to the highly conserved -YENPTY- motif in the APP C-terminus to regulate its trafficking, processing, and transcriptional modulation.27
To understand further the role of the Drosophila ortholog of APP-BP1 (dAPP-BP1), we have generated null mutations in dAPP-BP1. We have discovered that the loss of dAPP-BP1 blocks the NEDD8 conjugation pathway and causes apoptosis in imaginal disc cells. We find that dAPP-BP1 binds to the intracellular domain (ICD) of APPL, and dAPP-BP1 mutations rescue Appld phenotypes. Overexpression of APPL inhibits the NEDD8 conjugation pathway, disrupts the normal bristle pattern in the fly thorax, and induces apoptosis in wing imaginal discs. These APPL overexpression phenotypes are greatly enhanced by reduction in dAPP-BP1 level.
Results
dAPP-BP1 encodes the Drosophila homolog of APP-BP1, a component of the neddylation pathway
The dAPP-BP1LP67 mutant was identified in an enhancer-trap mutagenesis using the transposon P (w+lacZ) as mutagen. The screen selected for flies that were sluggish or died prematurely. Twenty mutant lines were identified: the dAPP-BP1LP67 mutation was one causing a shortened lifespan. The P-element insertion site for dAPP-BP1LP67 is located 79 bp upstream of the translation initiation site, within the 5′-untranslated region (UTR) of CG7828. CG7828 is homologous to human APP-BP1 (Figure 1c) and was named dAPP-BP1. Five cDNAs corresponding to dAPP-BP1 from the Berkeley Drosophila Genome Project were AT09990, AT15603, LP03107, AT26180, and BE977705. Sequence analysis showed that the most complete cDNA, AT09990, consists of 1846 bp, encoding a putative protein of 524 amino acids, with 165 bp of 5′UTR and 59 bp of 3′UTR. The dAPP-BP1 gene spans about 2.3 kb of genomic DNA. The gene contains six exons spliced to yield a single 1846 bp transcript, as determined by Northern blot analysis and cDNA sequencing (Figure 1b, data not shown). The deduced dAPP-BP1 shows canonical features present in human APP-BP1, including coiled coil motif and the UBA/THIF-type NAD/FAD-binding domain.
Molecular map and sequence alignment of dAPP-BP1. (a) Transposon insertion sites are indicated above the map by triangles: dAPP-BP1LP67 is located about 200 bp downstream of the transcription initiation site, within the 5′UTR of dAPP-BP1 mRNA. The dAPP-BP1Ex62 and dAPP-BP1Ex183 lines were obtained by P-element excision of dAPP-BP1LP67 and the excised regions are represented by horizontal arrows. In dAPP-BP1Ex62, 2789 bp is deleted: the first three exons and 30 bp of the fourth exon of dAPP-BP1. The dAPP-BP1Ex183 mutation has 1621 bp deleted. (b) Diagram of the dAPP-BP1 transcript. The 2.3 kb region indicated by a bracket shown in (a) is enlarged. The dAPP-BP1 gene has six exons and produces a 1846 bp transcript. (c) Sequence comparison of dAPP-BP1: dAPP-BP1 shares 36–43% identity with other APP-BP1 proteins from Arabidopsis to mammals
dAPP-BP1 is essential for Drosophila development
Two deletion mutants of dAPP-BP1 were generated by imprecise excision of the dAPP-BP1LP67 transposon. In dAPP-BP1Ex62 flies, 2789 bp was deleted, including the first three exons and 31 bases of the fourth exon of dAPP-BP1. The dAPP-BP1Ex183 mutation is a 1621 bp deletion including 84 bp of 5′UTR (Figure 1a). By polymerase chain reaction after reverse transcription of RNA (RT-PCR), we confirmed that dAPP-BP1Ex62 is a null mutant of dAPP-BP1 (Figure 2e). The dAPP-BP1Ex62 mutant is lethal in third instar larvae; lethality is rescued by overexpression of dAPP-BP1 (Supplementary Table S1). The temporal expression pattern of dAPP-BP1, examined using quantitative RT-PCR (Figure 2f), showed expression at all developmental stages.
dAPP-BP1 mutant phenotypes and developmental expression. (a) The adult eyes of w ey-flp;; P{Ubi-GFP}61EF P{neoFRT}80B/dAPP-BP1Ex183P{neoFRT}80B. (b) The adult eyes of w ey-flp;; P{Ubi-GFP}61EF P{neoFRT}80B/dAPP-BP1Ex62P{neoFRT}80B. The dAPP-BP1Ex183 clones containing white ommatidia exist in (a), compared to (b) in which dAPP-BP1Ex62 clones (marked by white ommatidia) are absent, leaving a defective eye that contains dAPP-BP1Ex62/+ or +/+ cells in red ommatidia. (c) Wild-type brain and eye imaginal disc. (d) dAPP-BP1Ex62/Df (3L) lxd6 eye discs are almost demolished. (e) mRNA expression analysis in third instar larvae (RT-PCR): dAPP-BP1 mRNA is not expressed in dAPP-BP1Ex62 homozygotes. (f) Quantitative RT-PCR analysis of dAPP-BP1 expression during development. As control, transcripts of rp49 were amplified. dAPP-BP1 transcripts can be detected during all developmental stages
Loss-of-function phenotypes for dAPP-BP1 mutations were examined by mosaic analysis. For dAPP-BP1Ex62 very few mutant cells could be found in adult eyes (Figure 2b). For dAPP-BP1Ex183 mosaics, however, mutant and wild-type clones were frequently observed (Figure 2a). Mutant clones were substantially smaller than wild type, suggesting that mutant cells were defective in proliferation or survival. Consistent with this interpretation, third instar larvae for dAPP-BP1Ex62 homozygote and dAPP-BP1Ex62/Df(3L)lxd6 genotypes have unusually small imaginal discs (Figure 2d). Taken together, these observations indicate that dAPP-BP1 is essential for normal tissue development in Drosophila.
The dAPP-BP1 mutation blocks the neddylation pathway
The APP-BP1 and UBA3 heterodimer has been reported to be responsible for E1-like functions in NEDD8 activation.2 If dAPP-BP1 participates in neddylation, mutant phenotypes would be expected to be similar to those of NEDD8 mutants. NEDD8 modification is necessary for SCFslimb-mediated protein degradation in Drosophila wing discs.13 Thus, protein stability was examined for substrates of Slimb in dAPP-BP1 mutant cells. In developing wing discs, dAPP-BP1 mutant cells accumulated high levels of full-length Ci (CiFL) and Arm proteins (Figure 3j–l and m–o). These are similar to accumulations previously reported for NEDD8 mutants.13
dAPP-BP1 mutation blocks NEDD8 conjugation pathway. Accumulation of CiFL, Armadillo, and Cul1 in dAPP-BP1 mutant cells. (a–c) Clones carrying dAPP-BP1Ex183 are marked by the lack of GFP expression (a, c). (a–i) The blue arrowheads mark the position of the morphogenetic furrow (MF) in this and following figures. For imaginal discs, anterior is to the left. (d–l) Clones carrying dAPP-BP1Ex62 are marked by the lack of GFP (d, g) or beta-galactosidase (j, m) expression. (d–f) A late third-instar eye disc containing dAPP-BP1Ex62 mutant clones and stained by anti-CiFL antibody. In mutant cells, CiFL protein levels (arrows) are similar in the MF and in the cells anterior to it. In posterior cells, CiFL persists, albeit at a lower level (arrowheads), and is not detected in the most posterior cells (asterisks). This pattern is similar to those observed in NEDD8 null clones.13 (g–i) A late third-instar eye disc containing dAPP-BP1Ex62 mutant clones and stained with anti-Cul1 antibodies (red). Cul1 accumulates in dAPP-BP1Ex62 mutant clones (arrows) that are marked by a lack of GFP expression (green in g, i). (j–l) Accumulation of Armadillo observed in dAPP-BP1Ex62 mutant clones in the wing disc (arrows). (m–o) Accumulation of CiFL observed in the dAPP-BP1Ex62 mutant clones in wig disc (arrows). (p) Ex62 is dAPP-BP1Ex62 homozygote cell extracts. Western blot analysis of cell extracts from third-instar eye discs and brain lobes by anti-Cul1 antibodies indicates that the Cul1 protein is accumulated and the neddylated form of Cul1 (arrow) is almost missing in the dAPP-BP1Ex62 homozygote
In eye discs, CiFL accumulated in dAPP-BP1 clones located anterior to the morphogenic furrow (MF) (Figure 3d–f, arrows). This is similar to CiFL accumulation observed in MF cells that transduce the Hh signaling pathway. CiFL accumulation was also observed in some posterior mutant cells. Similar to accumulation previously described for NEDD8 mutant clones,13 the CiFL accumulation observed here was in cells located proximal to the MF (Figure 3d–f, arrowheads), but not in cells located distal to it (Figure 3d–f, asterisks). Eye discs were also examined for accumulation of Cul1, since previous investigations13 have shown that neddylation controls Cul1 stability. Cul1 has been found to accumulate in NEDD8 mutant clones. Here, Cul1 was found to accumulate in dAPP-BP1 null clones (Figure 3g–i). Clones of dAPP-BP1Ex183 resembled wild-type clones (Figure 1a–c and data not shown). Neddylated forms of Cul1 are missing and unneddylated Cul1 is greatly accumulated in cell extracts from eye discs and larval brain (Figure 3p). Taken together, all of these observations appear to be due to loss of dAPP-BP1, with the indication that dAPP-BP1 is an essential component of the Drosophila neddylation pathway.
The loss of dAPP-BP1 causes apoptosis, not cell proliferation
Growth properties of dAPP-BP1 mutant cells were examined by comparing marked pairs (twin spots) of clones in the wing disc. Homozygous dAPP-BP1Ex62 mutant clones were found to be substantially smaller than their wild-type twins, by about a factor of three (Figure 4s and t). In addition, the dAPP-BP1Ex62 clones completely lacked elav-staining, indicating the absence of differentiated neurons. Interestingly, homozygous dAPP-BP1Ex183 mutant clones were found to be similar in size to their wild-type twins (Figure 4q and r). These observations indicate that the absence of dAPP-BP1 protein causes a decrease in growth during development.
The loss of dAPP-BP1 causes apoptosis, not cell proliferation. (a–d) Third instar eye discs. Scale bar is 50 μm. (a, b) TUNEL assay. White indicates TUNEL signals. (c, d) Immunostaining by anti-activated caspase 3. (Casp3=anti activated caspase 3). White indicates anti-activated caspase 3 positive site. (a) dAPP-BP1Ex62 homozygote. The size of eye disc is greatly reduced and TUNEL signals are greatly increased compared with the w1118 wildtype eye disc (b). (c) dAPP-BP1Ex62 homozygote. The level of activated caspase 3 is greatly increased in the wildtype, w1118 (d). Clones carrying dAPP-BP1Ex62 are marked by the lack of GFP (e, g, k, m) or beta-galactosidase (h, j, k, m) expression. (e–g) The level of activated caspase is increased in dAPP-BP1Ex62 clones in 3rd larvae eye disc (arrows). (h–j) The amount of activated caspase is increased in mutant clones in 3rd larvae wing disc (arrows). (k–m) PH3 is anti –phospho histone H3. dAPP-BP1Ex62 cells retain the ability to proliferate. (n–p) Clones carrying dAPP-BP1Ex62 are marked by the lack of myc expression. The number of elav-positive cells is decreased in dAPP-BP1Ex62 clones (arrows) (q) dAPP-BP1Ex183 homozygous clones and twin spots (white asterisk=dAPP-BP1Ex183 clones (arrows) homozygous clones were marked by lack of β-galactosidase expression. Black asterisk=twin spots. β-galactosidase=Green). (r) Area measurements of 31 pairs of homozygous dAPP-BP1Ex183 mutant and wild-type twin clones (scale: 100 pixels). Wild-type and mutant cells are produced in the same cell division, so differences in clone area reflect differences in growth or cell survival after clone induction. (s) dAPP-BP1Ex62 homozygous clones and twin spots (white asterisk=dAPP-BP1Ex62 homozygous clones were marked by lack of β-galactosidase expression. Black asterisk=twin spots. β-galactosidase=Green). (t) Area measurements of 42 pairs of homozygous dAPP-BP1Ex62 mutant and wild-type twin spots (scale: 100 pixels). The area of wild-type twin clones is about three times larger than that of mutant clones
Two possible mechanisms were considered, and experiments designed to distinguish between them. The first mechanism is that the absence of dAPP-BP1 could reduce growth via decreased cell proliferation. Alternatively, the absence of dAPP-BP1 could trigger apoptosis that would reduce growth through the removal of neuronal cells. Apoptosis was examined in the eye discs of dAPP-BP1Ex62 homozygotes. In normal eye discs, apoptosis is very low, as indicated by terminal deoxynucleotidyl transferase mediated-biotinylated UTP nick end labeling (TUNEL) markers and the cleaved (active) form of caspase 3 (Figure 4b and d). In contrast, there are massive levels of caspase 3 (Figure 4c) and TUNEL signal (Figure 4a) observed all across the eye discs of dAPP-BP1 mutants. Caspase 3 is also increased in dAPP-BP1Ex62 clones in wing discs and in eye discs (Figure 4e–j). The rate of cell division in dAPP-BP1 mutant clones was examined using an antibody to the phosphorylated form of histone 3 (PH3). PH3 staining is uniform over eye discs containing dAPP-BP1Ex62 clones, indicating that dAPP-BP1 null cells retain the ability to proliferate. Thus, these observations are consistent with a role for dAPP-BP1 in either the regulation of apoptosis or in the regulation of developmental pathways that, when improperly controlled, lead to apoptosis.
The dAPP-BP1 protein interacts with the amyloid precursor protein-like protein
Original identification of APP-BP1 was based on its binding with the ICD of APP.1 The binding site has been identified as the carboxyl-terminal 31 amino-acid residues of APP.4 This suggests the possibility of a similar interaction between dAPP-BP1 and APPL. Interaction was examined by an in vitro binding assay using a bacterially expressed GST-tagged APPLC63 (APPL carboxyl-terminal 63 amino-acid residues), GST, and in vitro expressed 35S-methionine-labeled dAPP-BP1. Using this assay, dAPP-BP1 protein is detected only in GST-APPLC63 pre-adsorbed glutathione-agarose beads (Figure 5f). Thus, similar to the described APP-BP1 interaction with APP, interaction is observed here between dAPP-BP1 and APPL via the APPL ICD.
dAPP-BP1 interacts with Appl. (a) Percent of normal scutellar macrochaete: Appld=Appld, Appld;183=Appld;; dAPP-BP1Ex183/+, Appld;62=Appld;; dAPP-BP1Ex62/+, OR=Oregon R. Missing macrochaete phenotype of Appld flies is suppressed by dAPP-BP1Ex62. (a–e) All of these flies were incubated at 25°C. (b–e) Fly scutellum. White circle indicates macrochaete. (b) Appld, (c) Appld;; dAPP-BP1Ex183/+, (d) Appld;; dAPP-BP1Ex62/+, and (e) Oregon R. (f) Binding studies performed with the bacterially expressed GST-tagged APPL C63 (APPL carboxy-terminal 63 amino acids), GST, and the in vitro-expressed dAPP-BP1. GST-APPL C63 specifically binds with dAPP-BP1. (g) Western blot analysis of cell extract from third-instar eye discs and brain lobes by anti-APPL antibodies. Loss of dAPP-BP1 slightly decreases APPL protein levels, but overexpression of dAPP-BP1 greatly increases the amount of APPL proteins. Da-GAL4 is expressed in all imaginal discs. (h–j) Adult thorax of flies incubated at 29°C, the temperature at which GAL4 induction is most efficient. White circle indicates macrochaete. (h) ap-GAL4/+, (i) UAS-dAPPBP1/+, (j) ap-GAL4/+; UAS-dAPPBP1/+. In the dAPP-BP1 overexpression condition, the size of the scutellum (arrows) is decreased and most macrochaetes are missing. (k) Western blot using an anti-dAPPBP1 antiserum. The amount of dAPP-BP1 is increased in Appld adult heads
Interaction between APPL and dAPP-BP1 was examined further by Western blot using anti-APPL antibody. This analysis shows that the level of APPL protein is correlated with the overall amount of dAPP-BP1. Cell extracts from a genetic background where dAPP-BP1 is overexpressed show that APPL levels are greatly increased (about six-fold) compared to wild-type controls. In a dAPP-BP1Ex62 mutant background, APPL levels are greatly reduced compared to wild-type controls.
The dAPP-BP1 mutation partially suppresses the thorax phenotype of Appld
In addition to physical interactions between dAPP-BP1 and APPL proteins, genetic interactions are observed between dAPP-BP1 and APPLd mutations. Previously, Appl mutations have been found to cause macrochaete deletions in the scutellar region of the adult thorax.28 The Appld macrochaete-missing phenotype is due to the deletion of external cells and includes the absence of macrochaete-associated neurons.28 Appld flies that are also heterozygous for dAPP-BP1Ex62 show rescue of the macrochaete (Figure 5a–e). In the dAPP-BP1Ex62 heterozygotes, approximately 30% of the missing anterior scutellar bristles are restored to the Appld mutants (Figure 5a). Thus, dAPP-BP loss of function acts as a suppressor of Appld. Overexpression of dAPP-BP1 also shows macrochaete-missing phenotype, and the protein level of dAPP-BP1 is increased when APPL is absent (Figure 5j and k and Supplementary Figure S1). From these observations, the suggestion is that Appld mutant phenotypes are caused by abnormal accumulation of dAPP-BP1. The excess quantity of dAPP-BP1 can lead to low levels of apoptosis with loss of APPL-induced macrochaete elimination due to cell death (Figure 7t).
dAPP-BP1 overexpression rescues APPLsd-induced apoptosis. (a–h) Adult wing of Drosophila. Scale bar is 1 mm. (a–m) The flies were raised at 25°C. (a) Wild-type wing expressing only the GAL4 driver (MS1096/Y). (b) MS1096/Y;UAS-dAPPBP1/+. Same as (a). (c) MS1096/Y;UAS-Appls/+. No difference compared to (a). (d) MS1096/Y;UAS-Appl/+. (e) Wings overexpressing APPLsd using the MS1096 GAL4 driver (MS1096/Y;UAS-Applsd/+) are smaller than (a) and the vein pattern is broken. (f) Co-expression of dAPP-BP1 (MS1096/Y;UAS-Applsd/UAS-dAPPBP1) rescues wing phenotype of (e). (g) Wings overexpressing APPLsdΔIC using the MS1096 GAL4 driver (MS1096/Y;UAS-ApplsdΔIC/+) are smaller than (a) and vein pattern is broken. (h) MS1096/Y;UAS-ApplsdΔIC/UAS-dAPPBP1 similar to (g). (i) MS1096/Y;UAS-Appl/UAS-Appl. Increased level of APPL causes small wing and cross-vein missing phenotypes (arrows). (j) Magnification of (i). Arrows indicate missing cross-vein. (k) Reduced level of Cul1 (MS1096/Y;UAS-Appl/Cul1Ex) enhances APPL overexpression phenotypes. (l) Magnification of (k). Arrows indicate missing cross-vein. (m) Quantification of the missing inter-vein phenotypes. P35 suppress APPL-overexpression pheotypes (1XAPPL=MS1096/Y;UAS-Appl/+, 2XAPPL=MS1096/Y;UAS-Appl/UAS-Appl, 1XAPPL;Cul1Ex=MS1096/Y;UAS-Appl/Cul1Ex, 1XAPPL;UAS-p35=MS1096/Y;UAS-Appl/UAS-p35). (n) Schematic overview of mutated APPL molecules used. ECD=extracellular domain, TM=transmembrane, ICD=intracellular domain, SP=signaling peptide, C=cleavage site. (o–v) The flies were incubated at 29°C. (o–t) Wing discs labeled with anti-activated caspase 3 antibodies. (o) MS1096 control. (p) MS1096;UAS-ApplsdΔIC/+. The apoptosis level is enhanced. (q) MS1096;UAS-ApplsdΔIC/UAS-dAPPBP1 similar to (p). (r) MS1096;UAS-Applsd/+. Discs overexpressing APPLsd have a high level of cell death. (s) MS1096;UAS-Applsd/UAS-dAPPBP1. dAPP-BP1 overexpression rescues APPLsd-induced apoptosis. (t) MS1096; UAS-dAPPBP1/UAS-dAPPBP1. dAPP-BP1 overexpression induce low levels of apoptosis. (u, v)TUNEL assays in wing discs. (u) MS1096;UAS-Applsd/+. (v) MS1096;UAS- Applsd/UAS-dAPPBP1. The level of cell death is decreased compared to (u)
APPL-induced toxicity is enhanced by dAPP-BP1 mutation
Genetic interactions were investigated using an Appl transgene driven by apterous (ap)-GAL4. For flies in the ap-Appl strain, thorax phenotypes resembled those observed in wild-type flies (Figure 6a, first panel). Loss of one dose of the dAPP-BP1 gene in an ap-Appl background, however, caused loss of thoracic macrochaetes and a reduction in scutellem size (Figure 6a, second panel, arrows). This phenotype resembles that seen in animals with two-fold Appl overexpression (Figure 6a, third panel, arrows). The dAPP-BP1 interaction with ap-Appl is sensitive to temperature. When reared at 29°C, flies that are haplo-deficient for dAPP-BP1 in an ap-Appl background show an extreme phenotype: the loss of thoracic macrochaetes is massive and the scutellar size reduction is severe (Figure 6c).
APPL-induced toxicity is enhanced by dAPP-BP1 mutation. (a) Quantification of the phenotypes in the adult thorax for different genetic backgrounds. 1XAPPL=ap-GAL4,UAS-Appl/CyO; 1XAPPL;EX62=ap-GAL4,UAS-Appl/CyO; dAPP-BP1Ex62/+; 2XAPPL=ap-GAL4,UAS-Appl/UAS-Appl; 1XAPPL;Cul1Ex=ap-GAL4,UAS- Appl/Cul1Ex. Cul1Ex is a deletion mutant of the Cul1 gene. In some genetic backgrounds, the size of scutellum (arrows) is decreased. These flies were raised at 25°C. (a–d) White circle indicates macrochaete. (b–d) The adult nota of flies raised at 29°C. (b) ap-GAL4,UAS-Appl. (c) ap-GAL4,UAS-Appl; dAPP-BP1Ex62. Almost all macrochaetes are missing, and the size of the scutellum is highly reduced (arrows). (d) ap-GAL4,UAS-Appl/UAS-p35; dAPP-BP1Ex62/+. (c) Phenotypes are partially rescued. Macrochaetes are restored and the size of the scutellum is almost completely rescued. (e–g) A wing disc stained for activated caspase 3 (white). The larvae were raised at 29°C. (e) ap-GAL4; dAPP-BP1Ex62/+. (f) ap GAL4,UAS-Appl/UAS-Appl. Signals are increased over those seen in (e). (g) ap GAL4,UAS-Appl/UAS-Appl; dAPP-BP1Ex62/+. The activated caspase-positive cells are increased compared with (f). (h, i) TUNEL assay in wing discs. The larvae were incubated at 29°C. (h) ap GAL4,UAS-Appl/UAS-Appl. (i) ap GAL4,UAS-Appl/UAS-Appl; dAPP-BP1Ex62/+. Apoptosis is greatly enhanced compared to (h). (j) Quantification of TUNEL signals in wing discs. Error bars represent the 95% confidence interval (2XAPPL=ap GAL4, UAS-Appl/UAS-Appl, 2XAPPL; Ex62=ap GAL4,UAS-Appl/UAS-Appl; dAPP-BP1Ex62/+). (k) APPL expression pattern in wing disc of ap GAL4, UAS-Appl/UAS-Appl
Genetic interactions between ap-Appl and Cul1 indicate that reduction in Cul1 level enhances ap-Appl thorax phenotypes (Figure 6a, fourth panel, arrows). This suggests that the neddylation pathway plays a role in the Appl-induced phenotypes observed in the thorax: neddylation is an essential feature of Cul1 functions. If these phenotypes are caused by apoptosis, overexpression of baculoviral P35 protein should rescue APPL-induced thorax phenotypes. Baculoviral P35 protein inhibits caspases, thereby blocking apoptosis in many species. Figure 6d shows that P35 rescues APPL-induced thorax phenotypes. The results of TUNEL and activated caspase 3 antibody staining in wing discs (Figure 6e–i) showed that APPL overexpression promoted apoptosis and this is enhanced by reduction of dAPP-BP1. The graph of Figure 6j confirms these results.
dAPP-BP1 overexpression suppresses APPLsd-induced apoptosis via the ICD of APPL
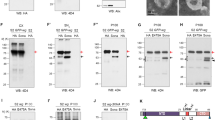
Ectopic expression phenotypes of APPL were examined using a GAL4 transgene to drive overexpression in the wing disc. MS1096 is an X-linked transgene that expresses GAL4 strongly on the dorsal surface of the wing primordium in third instar larvae; weak expression is observed on the ventral surface. Owing to dosage compensation, GAL4 levels in males (MS1096/Y) are twice those in heterozygous females (MS1096/+). APPLsd and APPLsdΔIC overexpression caused a great reduction in wing size for males (Figure 7e and g). Overexpression of APPLs and dAPP-BP1 caused minimal changes in the size of wings; wing size (Figure 7b and c) is nearly the same as for control flies (Figure 7a). In the case of APPL, one copy of Appl (Figure 7d) failed to show significantly different phenotypes. Increasing expression further using two copies of the UAS- Appl transgene reduced wing size and caused the loss of cross-veins (Figure 7i and j). The loss of cross-vein phenotype is rescued by co-expression of P35 (Figure 7m).
The reduction in wing size in APPL and its overexpression mutant forms could either be due to smaller cells or fewer cells. Every cell on the wing blade has a hair, so hair density marks cell density, and hence cell size. Wing hair density was unchanged on all the various APPL forms (data not shown). Therefore, in APPL and its mutant forms, overexpression causes a reduction in cell number responsible for the reduction of wing size. Apoptosis in wing imaginal discs was examined by TUNEL assay and immunostaining by anti-activated caspase 3 antibody. These experiments indicated a large increase in cell death throughout the wing pouch when APPLsd and APPLsdΔIC were overexpressed (Figure 7p, r and u). To test whether overexpression was sensitive to the dosage of dAPP-BP1, it was co-expressed with the APPL mutant forms. Only APPLsd-induced apoptosis and small-wing phenotypes were rescued by dAPP-BP1 overexpression (Figure 7f, h, q, s and v). When dAPP-BP1 was overexpressed alone, low levels of apoptosis occurred (Figure 7t). These data suggest that APPL-induced toxicity is attenuated by dAPP-BP1 via the APPL ICD.
Overexpression of APPL and its derivatives inhibits SCF ligase function
The Cul1Ex null mutation13 was used to examine interaction between APPL and SCF ligase. Hériché et al.29 reported that reduction of Cul1-mediated SCF function causes apoptosis. Here we show that reduction of Cul1 level enhances Appl-induced small wing and missing crossvein phenotypes (Figure 7k–m). Similarly, these phenotypes are enhanced by increasing the level of Appl (Figure 7i, j and m). To assess Cul1 function directly, we sought a target protein whose destruction might be limited to the Cul1-SCF/proteasome pathway. Ci levels are particularly sensitive to activity in the SCF/proteasome pathway. The level of Ci processing can be scored by measuring the accumulation of C-terminal epitope.29 This assay showed that reduction of Slimb and Roc1, that contribute to SCF function, is associated with increased Ci staining. Here, reduction of Cul1 function by overexpressing APPL and its mutant forms (Figure 8a–d) led to Ci accumulation. These results indicate that APPL and its overexpression mutant forms reduce Cul1-mediated SCF function.
Overexpression of APPL and its derivatives inhibits SCF ligase functions by the neddylation pathway. (a–d) Anti-CiFL staining in the third instar larval wing disc imaged with identical confocal microscope settings. All larvae were raised at 29°C. (a) Full-length Ci level in MS1096. (b) Increased level of full-length Ci in MS1096; UAS-Appl/UAS-Appl (arrow). (c) MS1096; UAS-Applsd/+. CiFL staining pattern is broadened and wing disc structure is abnormal. (d) MS1096; UAS-ApplsdΔIC/+ shows phenotype similar to (c). (e) The neddylation of Cul1 is repressed by overexpression of APPL or APPL derivatives. APPL, APPLsd, and APPLsdΔIC were overexpressed by Appl-GAL4 at 29°C. Black-lined box indicates neddylated form of Cul1 protein. (f–n) Third instar larval wing disc incubated at 25°C. The cell death induced by APPLsd and APPLsdΔIC is enhanced by dAPP-BP1 mutation. The amount of apoptosis is more severe when APPLsd is expressed. (f–j) Immunostaining by anti-active caspase 3 (white). (f) ap-GAL4; dAPP-BP1Ex62/+. (g) ap GAL4/UAS-ApplsdΔIC. (h) ap GAL4/UAS-ApplsdΔIC; dAPP-BP1Ex62/+. (i) ap GAL4/UAS- Applsd. (j) ap GAL4/UAS-Applsd; dAPP-BP1Ex62/+. (k–n) TUNEL assay in wing discs. (k) ap GAL4/UAS-ApplsdΔIC. (l) ap GAL4/UAS-ApplsdΔIC; dAPP-BP1Ex62. (m) ap GAL4/UAS-Applsd. (n) ap GAL4/UAS-Applsd; dAPP-BP1Ex62/+
To test if inhibition is related to neddylation, Western blot analysis was performed with anti-Cul1 antibody. APPL and its derivatives were overexpressed by Appl-GAL4 at 29°C. Cell extracts from third instar larval brain and eye discs were used for Western blots. Neddylated Cul1 is decreased in these cell extracts, with especially significant reductions observed for APPLsd. These results demonstrate that reduction of SCF function is due to inhibition of neddylation. Although dAPP-BP1 cannot suppress APPLsdΔIC-induced apoptosis, reduction of dAPP-BP1 enhances APPLsdΔIC-induced apoptosis (Figure 8g, h, k and l). This suggests that APPLsdΔIC-induced apoptosis is related to neddylation. For APPLsdΔIC and APPLsd levels of apoptosis are greater than for APPL. For APPL, increased cell death is observed only when two copies of APPL are overexpressed and flies are reared at 29°C (data not shown, Figure 6e–j). Because expression levels of these UAS lines are not significantly different,23 these results demonstrate that cleavage of APPL is important for its regulation. Overexpression of APPL consistently failed to produce abnormal phenotypes (Figure 7c).
Discussion
In this work, dAPP-BP1 mutants were identified, isolated, and characterized. Immunostaining and immunoblotting of the developing discs showed that loss of dAPP-BP1 blocks the neddylation pathway, suggesting that the role of dAPP-BP1 is similar to that of its mammalian counterpart. A loss-of-function mutant of dAPP-BP1 showed that mutant cells lacking dAPP-BP1 activity display elevated levels of apoptosis, but no changes in cell proliferation. Imaginal discs are of small size in the dAPP-BP1 null mutant, suggesting that apoptosis plays a significant role in reducing cell number. In dAPP-BP1 null clones, high levels of apoptosis are correlated with loss of photoreceptor neurons in the imaginal eye discs of mutants. Loss of dAPP-BP1 also hinders cell survival in other cell types. Our loss-of-function genetic analysis is in agreement with previous studies showing that mice deficient in the Uba3 gene that encodes a catalytic subunit of NEDD8-activating enzyme died in utero at the peri-implantation stage, with mutant embryos showing apoptosis of their inner cell mass.30
Like mammalian APP-BP1, dAPP-BP1 physically interacts with APPL ICD and dAPP-BP1 mutants genetically interact with Appld. The presence of dAPP-BP1 is necessary for attenuated Appl-induced toxicity via the ICD of APPL. The human APP-BP1-binding site is covered with the major APP carboxy-terminal adaptor protein-binding region.27 Several mammalian adaptor proteins are conserved in Drosophila. Binding by dAPP-BP1 may regulate an interaction between APPL ICD and other adaptor proteins. Physical interactions between dAPP-BP1 and APPL may be essential for dAPP-BP1-mediated apoptosis suppression, and this suppression results from APPL ICD processing or from signal transduction regulation. Chen et al.4 demonstrated that neurons expressing APP or APP (V642I) show an increase in APP-BP1 levels in lipid rafts, which is also observed in the hippocampal region of AD patients. This could explain why an elevated level of membrane APP increases the physical interaction between APP and APP-BP1 required for reducing APP-induced toxicity.
The major target of neddylation is the Cullin family of proteins.7 Our study indicates that the membrane-bound form of APPL inhibits SCF ligase functions. Previous studies indicated that the loss of Cul1-mediated SCFslimb ligase function causes apoptosis and DNA synthesis by abnormal accumulation of E2F.29 APP overexpression also causes DNA synthesis and apoptosis in neurons.4 Since dAPP-BP1 directly binds to APPL ICD and the mutation suppresses Appld phenotypes, we suggest that Appl-induced SCF ligase inhibition is due to changes in the neddylation level of Cullin. Supporting this, Western blot analysis reveals that neddylated Cul1 level is reduced by overexpression of APPL. Also, dAPP-BP1 protein accumulates in Appld flies and overexpression of dAPP-BP1 causes a macrochaete-missing phenotype. If APPL is missing, dAPP-BP1 is hyperactivated and the neddylation level of Cullin is increased. When APPL is overexpressed, neddylation is decreased. Increased levels of neddylated Cullin do not necessarily mean enhanced SCF activity. Since CSN (COP9 signalosome) has deneddylase activity, the neddylated Cullin level is greatly increased in CSN mutant cells.31 SCF ligase substrates accumulate under these conditions.32 Interaction between Cul1 and p120CAND1 is required for SCF function,33 but p120CAND1 binds only to unneddylated Cul1.11, 34 For this reason, proper levels of neddylation may be important for SCF function.
Some APPL overexpression phenotypes are similar to Cullin mutant phenotypes. Torroja et al.23 reported that overexpression of wild-type and mutant forms of APPL in Drosophila motor neurons caused a dramatic increase in synaptic bouton number. Interestingly, recent studies show that synaptic bouton number in motor neuron endings was greatly increased in a Drosophila Cul-5 mutant.35 APPLsd and APPLsdΔIC overexpression also caused abnormalities in the adult mushroom bodies, with the lobes being less uniform and having fuzzy boundaries.26 APPL overexpression has been shown to induce post-developmental neurite arborization.36 Similarly, Zhu et al.37 showed the cell-autonomous involvement of Drosophila Cul3 in axonal arborization and dendritic elaboration of mushroom body neurons. The neddylation of cullins is important for their proper functioning, so that the APPL overexpression phenotypes of these reports may be directly related to reduced neddylation.
The extracellular region of APPL can partially inhibit the neddylation pathway when the transmembrane domain is intact. This repression is enhanced by the ICD. The extracellular domain of APP cannot interact physically with APP-BP1; consequently, it is possible that dAPP-BP1 is not the target of APPL-induced inhibition. However, the close correlation between their respective levels suggests that APPL could be targeting dAPP-BP1. We propose two modes of APPL regulation in Drosophila development (Figure 9). If APPL is cleaved at the extracellular cleavage site, it does not cause apoptosis. When the membrane-bound form of APPL increases, it interacts antagonistically with dAPP-BP1 and/or other neddylation proteins. As a result of this repression, the level of neddylated cullins, SCF ligase activity, and other cullin-mediated ligase activities are all reduced. The reduction of ligase activity then causes apoptosis. Direct interaction between dAPP-BP1 and APPL ICD can attenuate membrane-bound APPL functions. Mechanisms underlying the APP pathway are closely conserved in Drosophila and in mammals. Consequently, the study of Drosophila Appl and dAPP-BP1 can contribute substantially to the elucidation of APP function.
Materials and Methods
Immunocytochemistry of larval discs
Staged larvae are dissected in 1 × PBS. Dissected head cuticles are incubated in fixation solution (0.1 M PIPES, pH 6.9, 1 mM EDTA, 1.0% Triton X-100, 2 mM MgSO4, 1% EM grade formaldehyde) for 30 min. After incubating in blocking solution (50 mM Tris-Cl, pH 6.8, 150 mM NaCl, 0.5% NP-40, 5 mg/ml BSA, 0.01% Thimerosal) at 4°C for 1 h, head cuticles are transferred to each primary antibody in wash solution (50 mM Tris-Cl, pH 6.8, 150 mM NaCl, 0.5% NP-40, 1 mg/ml BSA, 0.01% Thimerosal) and incubated at 4°C overnight. After washing four times in wash solution, head cuticles are incubated with secondary antibody at room temperature for 2 h. After washing three times with wash solution, head cuticles are dissected in antifade solution. Primary antibodies were mouse anti-Armadillo (N27A1, 1:50, Iowa Hybridoma Bank), rat anti-elav (7E8A10, 1:500, Iowa Hybridoma Bank), rabbit anti-β-gal (1:1000, Sigma), mouse anti-GFP (1:200, Sigma), rat anti-Ci (2A1, 1:3),38 rabbit anti-Cul1 (1:250, Zymed), rabbit anti-cleaved (activated) caspase 3 (1:50, Cell Signaling Technology), rabbit anti-phospho-histone H3 (1:500, Upstate Biotechnology), rabbit anti-APPL (Ab952, 1:500),21 and mouse anti-Myc (1:200, Roche). Secondary antibodies coupled to the appropriate fluorophore (FITC, Cy3, or Cy5; Jackson Immunoresearch Laboratories) were diluted to 1:200 in wash solution. All confocal images were taken on an LSM510 (Carl Zeiss). Images were processed in Adobe Photoshop.
DNA constructs for germline transformation
Wild-type dAPP-BP1 full-length cDNAs were cloned into the pUAST vector and pUAST-HA vector as EcoRI–XbaI fragments. P-element-mediated germline transformation was performed according to described methods.39 The parental strain for all germline transformations was w1118. Flies bearing autosomal transgenes were identified and used for all analyses.
Fly strains
All stocks were maintained and raised under standard conditions. Oregon R wild-type and w1118 were used as control strains. Appld, Appl-GAL4, UAS-Appl+, Applsd, and Appls were gifts from Kalpana White22 and ap-GAL4 was a gift from P Adler. UAS-ApplsdΔIC was provided by Renato Paro. The MS1096 stock is as described in Capdevila and Guerrero.40 The Cul1 allele Cul1Ex was described previously.13 The Bloomington Stock Center (Bloomington, IN; http://flystocks.bioindiana.edu/) provided other fly stocks.
TUNEL assay
TUNEL assays were performed using the Roche in situ cell death detection kit (fluorescein, Catalog number 1 684 795). In brief, imaginal discs were fixed with fixation solution (0.1 M PIPES, pH 6.9, 1 mM EDTA, 1.0% Triton X-100, 2 mM MgSO4, 1% EM grade formaldehyde) for 30 min and washed with PBT (PBS, 0.1% Triton X-100) four times for 10 min. Discs were then washed twice for 5 min with PBT 5 × (PBS, 0.5% Triton X-100) and transferred to a reaction buffer for 10 min, followed by working-strength reaction buffer containing terminal deoxynucleotide transferase. The discs were incubated with the enzyme in a humidified chamber at 37°C for 2 h. The reaction was stopped by transferring the discs into PBT 5 × and discs were then rinsed in three changes of PBT. Discs were then immunoreacted with antibodies or directly imaged by confocal microscopy.
Clonal analysis
Mitotic recombination clones were induced 48±3 h after egg laying (AEL) in staged larvae by heat shock at 37°C for 80 min. Larval genotypes were: P{hsFLP}22 y1w*; P{arm-lacZ.V} 70C P{neoFRT} 80B/dAPPBP1ex62 P{neo FRT} 80B (or P{arm-lacZ.V} 70C P{neoFRT}80B/dAPPBP1ex183P{neoFRT} 80B). Discs were dissected at 110±3 h AEL, fixed in fixation solution, and stained with anti-β-galactosidase antibody to mark the clones. The following flies were used to generate dAPP-BP1 clones in the eye disc: yd2w1118P{ey-FLP.N}2; P{Ubi-GFP} 61EF P{neoFRT}80B/dAPPBP1Ex62 P{neoFRT}80B (or P{Ubi-GFP}61EF P{neoFRT}80B/dAPPBP1Ex183P{neoFRT}80B) and yd2w1118P{ey-FLP.N}2; P{πM}75C P {neoFRT} 80B/dAPP-BP1Ex62 P{neoFRT}80B (or P{πM}75C P{neoFRT}80B/dAPPBP1Ex183P{neoFRT}80B). Clone areas were measured using Adobe Photoshop 6.0.
Western blotting and antibody generation
A dAPP-BP1 cDNA (corresponding to amino-acid residues 147–299) was cloned into pET-15b vector that encodes a fusion protein with 6 × His tags. The fusion protein was expressed in Escherichia coli and purified with His·Bind Resin (Novagen). Polyclonal antibodies were produced by immunizing guinea pigs using the purified fusion protein as antigen (KOMA Biotech, Korea).
Larval brain and eye discs or adult heads were collected in 2 × SDS sample buffer and boiled for 10 min. The extracts were then run on a 3–8% Tris-acetate gel (Invitrogen) or 12% Tris-glycine gel. Western blotting was performed according to standard protocol. Primary antibodies used were rabbit anti-Cul1 (1:750; Zymed), rabbit anti-APPL (1:4000; gift from K White), guinea-pig anti-dAPP-BP1 (1:500), and β-tubulin (E7, Iowa Hybridoma Bank, 1:2000). After incubation with HRP-coupled secondary antibodies (goat anti-rat diluted 1:3000 and goat anti-rabbit diluted 1:3000; Pierce), blots were visualized using a chemiluminescent detection kit (ECL; Amersham Biosciences).
RT-PCR and quantitative RT-PCR
For RT-PCR, total RNA was prepared using TRIzol (Invitrogen) and reverse transcribed using M-MLV reverse transcriptase (Promega). PCR was performed using dAPP-BP1 specific primer including an intron with forward sequence 5′-CATATGCGGTTATGGGAGCTTAAC-3′ and reverse sequence 5′-GGATCCCTGCTTAAGGTACTCCTG-3′ to obtain a product of 640 bp. The GAP-DH control was amplified with the forward primer 5′-GTCAA CGATCCCTTCATCGA-3′ and reverse primer 5′-TGTACGATAGTTGTGGCTAG-3′. For quantitative RT-PCR, total RNA was isolated from each stage of Oregon R. First-strand cDNA was synthesized from 3 μg of total RNA with a total cycle number of 25. For control, rp49 primers were used.
GST pull-down analysis
35S-labeled dAPP-BP1 was synthesized in vitro using pSP64-AT09990 (dAPP-BP1 expressed sequence tag (EST)), TNT Quick-Coupled Transcription/Translation system (Promega), and L-[35S]methionine (Amersham Biosciences) according to the manufacturer's protocol. BL21 cells containing GST and GST-APPL C63 (GST-tagged APPL, carboxy-terminal 63 amino-acid residues) expression plasmids (pGEX 6P-1) were grown in LB media. Expression of fusion proteins was induced by treating cells with 0.5 mM isopropyl-1-thio-beta-D-galactopyranoside for 12 h at 20°C. GST fusion protein was purified by the batch GST Purification Module (Amersham Biosciences). GST-APPL C63 fusion protein and GST were isolated by using glutathione Sepharose 4 fast flow beads. The immobilized GST and GST-APPL C63 protein beads were washed with phosphate-buffered saline and incubated with 35S-labeled dAPP-BP1 at 4°C for 12 h in binding buffer (20 mM Hepes, 0.1 mM EDTA, 10 mM KCL, 10% glycerol, 0.4% Triton-X 100 and Sigma protease inhibitor cocktail). Beads were washed three times with buffer and subjected to SDS-polyacrylamide gel electrophoresis and autoradiography.
Abbreviations
- AD:
-
Alzheimer's disease
- ap :
-
apterous
- APP:
-
β-amyloid precursor protein
- APP-BP1:
-
β-amyloid precursor protein binding protein 1
- APPL:
-
APP-like protein
- Ci:
-
cubitus interruptus
- Da :
-
daughterless
- ECD:
-
extracellular domain
- EST:
-
expressed sequence tag
- ICD:
-
intracellular domain
- RT-PCR:
-
polymerase chain reaction after reverse transcription of RNA
- SDS-PAGE:
-
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- SCF complexes:
-
Roc1/Rbx1/Hrt1-Skp1-Cullin/Cdc53-F box complexes
- TUNEL:
-
terminal deoxynucleotidyl transferase mediated-biotinylated UTP nick end labeling
- UTR:
-
untranslated regions
References
Chow N, Korenberg JR, Chen X-N, Neve RL . APP-BP1, a novel protein that binds to the carboxyl-terminal region of the amyloid precursor protein. J Biol Chem 1996; 271: 11339–11346.
Hochstrasser M . Evolution and function of ubiquitin-like protein-conjugation systems. Nat Cell Biol 2000; 2: E153–E157.
Chen Y, McPhie DL, Hirschberg J, Neve RL . The amyloid precursor protein-binding protein APP-BP1 drives the cell cycle through the S–M checkpoint and causes apoptosis in neurons. J Biol Chem 2000; 275: 8829–8835.
Chen Y, Liu W, McPhie DL, Hassinger L, Neve RL . APP-BP1 mediates APP-induced apoptosis and DNA synthesis and is increased in Alzheimer's disease brain. J Cell Biol 2003; 163: 27–33.
Pan ZQ, Kentsis A, Dias DC, Yamoah K, Wu K . Nedd8 on cullin: building an expressway to protein destruction. Oncogene 2004; 23: 1985–1997.
Xirodimas DP, Saville MK, Bourdon JC, Hay RT, Lane DP . Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell 2004; 118: 83–97.
Deshaies RJ . SCF and Cullin/Ring H2-based ubiquitin ligases. Annu Rev Cell Dev Biol 1999; 15: 435–467.
Karin M, Ben-Neriah Y . Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity. Annu Rev Immunol 2000; 18: 621–663.
Kondo K, Kaelin JWG . The von Hippel–Lindau tumor suppressor gene. Exp Cell Res 2001; 264: 117–125.
Ohh M, Kim WY, Moslehi JJ, Chen Y, Chau V, Read MA et al. An intact NEDD8 pathway is required for Cullin-dependent ubiquitylation in mammalian cells. EMBO Rep 2002; 3: 177–182.
Liu J, Furukawa M, Matsumoto T, Xiong Y . NEDD8 modification of CUL1 dissociates p120CAND1, an inhibitor of CUL1-SKP1 binding and SCF ligases. Mol Cell 2002; 10: 1511–1518.
Wu K, Chen A, Tan P, Pan ZQ . The Nedd8-conjugated ROC1-CUL1 core ubiquitin ligase utilizes Nedd8 charged surface residues for efficient polyubiquitin chain assembly catalyzed by Cdc34. J Biol Chem 2002; 277: 516–527.
Ou CY, Lin YF, Chen YJ, Chien CT . Distinct protein degradation mechanisms mediated by Cul1 and Cul3 controlling Ci stability in Drosophila eye development. Genes Dev 2002; 16: 2403–2414.
Mori F, Nishie M, Piao YS, Kito K, Kamitani T, Takahashi H et al. Accumulation of NEDD8 in neuronal and glial inclusions of neurodegenerative disorders. Neuropathol Appl Neurobiol 2005; 31: 53–61.
Rosen DR, Martin-Morris L, Luo LQ, White K . A Drosophila gene encoding a protein resembling the human beta-amyloid protein precursor. Proc Natl Acad Sci USA 1989; 86: 2478–2482.
Kang J, Lemaire HG, Unterbeck A, Salbaum JM, Masters CL, Grzeschik KH et al. The precursor of Alzheimer's disease amyloid A4 protein resembles a cell-surface receptor. Nature 1987; 325: 733–736.
Luo LQ, Martin-Morris LE, White K . Identification, secretion, and neural expression of APPL, a Drosophila protein similar to human amyloid protein precursor. J Neurosci 1990; 10: 3849–3861.
Sisodia SS . Cellular and molecular genetic approaches in Alzheimer's disease. Neurobiol Aging 1994; 15: S135–S137.
Slunt HH, Thinakaran G, Von Koch C, Lo AC, Tanzi RE, Sisodia SS . Expression of a ubiquitous, cross-reactive homologue of the mouse beta-amyloid precursor protein (APP). J Biol Chem 1994; 269: 2637–2644.
Luo L, Tully T, White K . Human amyloid precursor protein ameliorates behavioral deficit of flies deleted for Appl gene. Neuron 1992; 9: 595–605.
Torroja L, Luo L, White K . APPL, the Drosophila member of the APP-family, exhibits differential trafficking and processing in CNS neurons. J Neurosci 1996; 16: 4638–4650.
Torroja L, Chu H, Kotovsky I, White K . Neuronal over-expression of APPL, the Drosophila homologue of the amyloid precursor protein (APP), disrupts axonal transport. Curr Biol 1999a; 9: 489–493.
Torroja L, Packard M, Gorczyca M, White K, Budnik V . The Drosophila beta-amyloid precursor protein homolog promotes synapse differentiation at the neuromuscular junction. J Neurosci 1999b; 15: 7793–7803.
Gunawardena S, Goldstein LSB . Disruption of axonal transport and neuronal viability by amyloid precursor protein mutations in Drosophila. Neuron 2001; 32: 389–401.
Tschape JA, Hammerschmied C, Muhlig-Versen M, Athenstaedt K, Daum G, Kretzschmar D . The neurodegeneration mutant lochrig interferes with cholesterol homeostasis and Appl processing. EMBO J 2002; 21: 6367–6376.
Li Y, Liu T, Peng Y, Yuan C, Guo A . Specific functions of Drosophila amyloid precursor-like protein in the development of nervous system and nonneural tissues. J Neurobiol 2004; 61: 343–358.
King GD, Scott Turner R . Adaptor protein interactions: modulators of amyloid precursor protein metabolism and Alzheimer's disease risk? Exp Neurol 2004; 185: 208–219.
Merdes G, Soba P, Loewer A, Bilic MV, Beyreuther K, Paro R . Interference of human and Drosophila APP and APP-like proteins with PNS development in Drosophila. EMBO J 2004; 23: 4082–4095.
Hériché JK, Ang D, Bier E, O'Farrell PH . Involvement of an SCFSlmb complex in timely elimination of E2F upon initiation of DNA replication in Drosophila. BMC Genet 2003; 4: 9–21.
Tateishi K, Omata M, Tanaka K, Chiba T . The NEDD8 system is essential for cell cycle progression and morphogenetic pathway in mice. J Cell Biol 2001; 155: 571–579.
Bech-Otschir D, Seeger M, Dubiel W . The COP9 signalosome: at the interface between signal transduction and ubiquitin-dependent proteolysis. J Cell Sci 2002; 115: 467–473.
Cope GA, Deshaies RJ . COP9 signalosome: a multifunctional regulator of SCF and other cullin-based ubiquitin ligases. Cell 2003; 114: 663–671.
Feng S, Shen Y, Sullivan JA, Rubio V, Xiong Y, Sun TP et al. Arabidopsis CAND1, an unmodified CUL1-interacting protein, is involved in multiple developmental pathways controlled by ubiquitin/proteasome-mediated protein degradation. Plant Cell 2004; 16: 1870–1882.
Zheng J, Yang X, Harrell JM, Ryzhikov S, Shim EH, Lykke-Andersen K et al. CAND1 binds to unneddylated CUL1 and regulates the formation of SCF ubiquitin E3 ligase complex. Mol Cell 2002; 10: 1519–1526.
Ayyub C, Sen A, Gonsalves F, Badrinath K, Bhandari P, Shashidhara LS et al. Cullin-5 plays multiple roles in cell fate specification and synapse formation during Drosophila development. Dev Dyn 2005; 232: 865–875.
Leyssen M, Ayaz D, Hebert SS, Reeve S, De Strooper B, Hassan BA . Amyloid precursor protein promotes post-developmental neurite arborization in the Drosophila brain. EMBO J 2005; 24: 2944–2955.
Zhu S, Perez R, Pan M, Lee T . Requirement of Cul3 for axonal arborization and dendritic elaboration in Drosophila mushroom body neurons. J Neurosci 2005; 25: 4189–4197.
Motzny CK, Holmgren R . The Drosophila cubitus interruptus protein and its role in the wingless and hedgehog signal transduction pathways. Mech Dev 1995; 1: 137–150.
Spradling A, Rubin GM . Genetic transformation of Drosophila with transposable element vectors. Science 1982; 4570: 348–353.
Capdevila J, Guerrero I . Targeted expression of the signaling molecule decapentaplegic induces pattern duplications and growth alterations in Drosophila wings. EMBO J 1994; 13: 4459–4468.
Acknowledgements
We thank Daria Hekmat-Scafe, Jeff Tan, Ed Glasscock, Shefa Gordon, and Juan Song for discussion and contributions to this work. The research was supported by a grant (M103KV010002 04K2201 00230) from Brain Research Center of the 21st Century Frontier Research Program funded by the Ministry of Science and Technology, the Republic of Korea to JY, and grants from the NIH and the Epilepsy Foundation to MT.
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by J Abrams
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Supplementary information
Rights and permissions
About this article
Cite this article
Kim, HJ., Kim, SH., Shim, SO. et al. Drosophila homolog of APP-BP1 (dAPP-BP1) interacts antagonistically with APPL during Drosophila development. Cell Death Differ 14, 103–115 (2007). https://doi.org/10.1038/sj.cdd.4401935
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.cdd.4401935
Keywords
This article is cited by
-
APLP1 promotes dFoxO-dependent cell death in Drosophila
Apoptosis (2015)
-
Analysis of amyloid precursor protein function in Drosophila melanogaster
Experimental Brain Research (2012)