Abstract
One notable phenotypic change during the differentiation of immature thymocytes into either mature CD4 or CD8 single-positive lineages is the acquisition of a resistance to glucocorticoid (GC)-induced apoptosis. We have previously reported that SRG3 is critical in determining the sensitivity for the GC-induced apoptosis in developing thymocytes. We report here that Notch signaling downregulates the transcriptional activation of SRG3 through N-box and/or E-box elements on its promoter. RBP-J represses SRG3 transcription through the N-box motif. On the other hand, Deltex1 competitively inhibits the binding of p300 to E2A/HEB protein bound to the E-box elements and represses the SRG3 promoter activity. Moreover, enforced expression of Deltex1 restored double-positive (DP) thymocyte survival from the GC-induced apoptosis. Our results suggest that Notch signaling confers differentiating DP thymocytes resistance to GCs by regulating the SRG3 expression through Deltex1, and that Deltex1 and SRG3 may play a significant role during DP thymocyte maturation.
Similar content being viewed by others
Introduction
Thymocytes differentiate from their bone marrow-derived precursors to mature cells through highly regulated processes. The differentiation of immature CD4+ CD8+ double-positive (DP) thymocytes into mature CD4 and CD8 single-positive (SP) thymocytes is critical for shaping mature T-cell repertoire and is exquisitely regulated. DP thymocytes are selected on the basis of their ability to recognize self-peptide/major histocompatibility complex (MHC) antigen by their T-cell receptors (TCRs).1, 2 Selected thymocytes further differentiate to become mature SP thymocytes and are self-MHC restricted in antigen recognition and tolerant to self-antigens.3, 4 Signaling through TCR, costimulatory molecules, and Notch1 is known to be important for selections and further differentiation of DP thymocytes into SP cells.5, 6, 7, 8 An interesting phenotypic change during the differentiation of DP thymocytes is the gaining of resistance to glucocorticoids (GCs). DP thymocytes are sensitive to GC-induced apoptosis, whereas SP thymocytes are relatively resistant to it.4, 9 Signaling through TCR and Notch1 confers resistance to GCs to developing thymocytes,10, 11, 12 suggesting a possibility that Notch signaling, together with TCR signaling, is involved in the maturation of DP thymocytes into SP stages.5
The molecular basis for the acquisition of GC resistance during maturation of DP thymocytes into SP cells has been implicated in recent studies.10, 11, 13 We have shown that the GC sensitivity is critically dependent on the expression level of SRG3.14 SRG3 is a mouse homolog of yeast SWI3 and human BAF155, a core component of SWI/SNF chromatin remodeling complex. It was initially isolated as a gene expressed at a high level in the thymus, but at a low level in the periphery.14 SRG3 deficiency leads to embryonic lethality like other components of the BAF complexes and its haploinsufficiency partly displays exencephaly.15 The ectopic expression of anti-sense SRG3 RNA in S49.1 thymoma cells reduces the GC sensitivity,14 while transgenic overexpression of SRG3 in peripheral T cells increase the susceptiblity to GC-induced apoptosis.16 We have shown that TCR signaling inhibits the SRG3 expression via Ras/MEK/ERK and PI3K pathways.17 Id3, an inhibitor of E2A, represses the SRG3 expression by preventing E2A/HEB complex from binding to the E-box element in the SRG3 promoter and, thereby, reduces the GC sensitivity in developing thymocytes.18 We have also reported that nitric oxide (NO) is involved in protecting immature thymocytes from the GC-induced apoptosis by repressing the SRG3 expression.19 These results strongly suggest that SRG3 is a critical determinant for the GC-induced apoptosis in thymocytes and that GC resistance is rendered by downregulation of the SRG3 expression via TCR-mediated signaling. In addition, Notch signaling also confers developing thymocytes resistance to GCs.10 Transgenic expression of SRG3 resulted in the restoration of GC sensitivity in thymocytes expressing transgenic Notch1, suggesting that Notch1 confers developing thymocytes resistance to GCs also by regulating the SRG3 expression.13 However, downstream modulators of Notch1 signaling regulating the GC sensitivity in developing thymocytes remain unknown.
Notch signaling regulates gene expression by activating either RBP-J (also termed CSL, CBF-1, Su(H), or Lag-1)6 or Deltex proteins.20 RBP-J requires the RAM and ankyrin domains of the activated form of Notch1 (NotchIC) for the transactivation of downstream target genes, whereas Deltex1 interacts with the first three ankyrin repeats of NotchIC.21 Downstream events of the Notch signaling pathway leading to GC resistance are suggested to be mediated by RBP-J.10 This is based on the observations that a Notch construct containing a deletion of the RAM domain and the first two ankyrin repeats (Notch/ANK) does not mediate the GC-induced apoptosis and that NotchIC with point mutations in the RAM domain inhibiting RBP-J interaction also shows similar effects, but at a reduced level.10 However, since Deltex1 interacts with the first three ankyrin repeats of NotchIC,21 the possibility that the downstream effects of the Notch signaling may also be mediated by the Deltex1-dependent mechanism cannot be completely excluded. Furthermore, the level of RBP-J expression is not significantly changed during thymocyte differentiation from double negative (DN) to DP and from DP to SP stages,22 whereas Deltex1 is expressed at a relatively high level in CD4 and CD8 SP thymocytes and at a low level in DP thymocytes.20, 22 Recently, it has been reported that RBP-J deficiency does not significantly perturb the development of DP thymocytes into CD4 and CD8 SP lineages.23 These results suggest a possibility that Deltex1 may be a major downstream modulator of the Notch signaling pathway controlling the maturation and the GC sensitivity in developing thymocytes.
Notch1 and Deltex1 have been reported to inhibit the transcriptional activity of bHLH transcription factor E2A.24, 25 It has been shown that the inhibition of transcriptional activity of bHLH MASH1 and HNF4 is mediated by the direct physical interaction between Deltex1 and coactivator p300.21 In addition, the interaction mechanism between E2A and p300/CBP is reported in insulin gene transcription and in pre-B cells, respectively.26, 27 E2A/HEB binding to the E-box element on SRG3 promoter regulates the SRG3 expression and, thereby, controls the sensitivity to GC-induced apoptosis in thymocytes.18 Therefore, it is possible that Notch-induced Deltex1 may inhibit the transcriptional activity of E2A by blocking the recruitment of a coactivator(s), and this will result in reduced expression of SRG3 and resistance to the GC-induced apoptosis in developing thymocytes.
We report here that Notch signaling downregulated the transcriptional activation of SRG3 through N-box and/or E-box elements on its promoter. RBP-J repressed the SRG3 transcription through the N-box motif. On the other hand, Deltex1 competitively inhibited the binding of p300 to E2A/HEB protein bound to the E-box elements and repressed the SRG3 promoter activity. The SRG3 promoter activity was restored from the Notch-mediated repression by the expression of dominant-negative form of Deltex1 (DN-Deltex1). Furthermore, enforced expression of Deltex1 restored DP thymocyte survival from the GC-induced apoptosis. Our results indicate that Deltex1 activated by Notch signaling may confer differentiating DP thymocytes resistance to GCs by regulating the SRG3 expression, and suggest that Deltex1 and SRG3 may play a significant role during thymocyte differentiation.
Results
Ankyrin repeat region of NotchIC is required for the Notch-mediated repression of SRG3
We have previously shown that NotchIC represses the promoter activity of the SRG3 gene.13 We then investigated what molecules are involved in the SRG3 repression in the downstream pathway of the Notch signaling. To examine the downstream pathways of Notch signaling repressing the SRG3 expression, several NotchIC derivatives (Figure 1a) were cotransfected into 16610D9 cells. Deletion of all the Notch derivatives was verified by sequencing and their expression in COS-1 cells was confirmed by immunoblotting with anti-Myc antibody (data not shown). It has been reported that RBP-J requires the RAM and ankyrin domains of NotchIC for the transactivation of downstream target genes, whereas Deltex1 interacts with the first three ankyrin repeats of Notch.21 The inhibitory effect on the SRG3 reporter was yielded only with the NotchIC/ANK construct lacking the RAM domains (maximum 66% repression), whereas ectopic expression of NotchIC/ANK2, deleted with the RAM domain and the first two ankyrin repeats, did not (Figure 1b). However, minor repression was seen with constructs that had complete deletion of the ankyrin repeats (maximum 22% repression). These results strongly suggest that the Notch1/Deltex1 signaling pathway, which requires the ankyrin repeats, acts as a major regulatory pathway, while it appears that Notch-mediated repression of the SRG3 expression may partially be dependent on Notch/RBP-J signaling pathway via the RAM domain.
Ankyrin repeat region of NotchIC is required for Notch-mediated repression of SRG3. (a) Schematic representation of various deletion mutants of NotchIC. NotchIC/ANK lacking the RAM domain begins at the first amino acid of the ankyrin repeats and NotchIC/ANK2 begins at the third ankyrin repeat of Notch1. NotchIC/dANK contains the RAM domain, but lacks all six ankyrin repeats. (b) Ankyrin repeats of NotchIC are required for the repression of SRG3 promoter activity. 16610D9 cells were transfected with 3 μg of D2 luciferase reporter construct containing the 1.1 kb promoter fragment of SRG317 and various amounts of Myc-tagged Notch1 deletion mutants as indicated in Materials and Methods. The results were plotted from three independent experiments. Error bars represent standard deviation
RBP-J and/or Deltex1 repress SRG3 transcription
It is known that Notch/RBP-J signaling pathway acts on the N-box consensus sequence, CACNAG, and Hes proteins bind directly to N-box sequence to repress the transcriptional activity of their target genes.28 The finding of one N-box motif in the SRG3 promoter by homology search prompted us to examine the functional role of RBP-J in regulating the SRG3 promoter activity. We found that overexpression of RBP-J repressed the luciferase reporter activity driven by 1.1 kb promoter fragment of SRG3 (D2) in the murine thymoma cell line, 16610D9 (approximately 52% reduction) (Figure 2b). Such repression requires an intact N-box, because the repression was absent when the N-box was mutated (D2/m745). These results indicate that RBP-J repressed the SRG3 expression through the N-box sequence in the promoter (Figure 2b).
RBP-J and/or Deltex1 repress SRG3 transcription. (a) Reporter constructs of the SRG3 promoter, wild-type proximal (D2), and minimal (D7) SRG3 promoters were described previously.13, 17 Closed circles represent one N-box and two E-box elements. The D7/m745 construct contains mutant N-box (CACCAG into CTCGAG, ○). (b) Overexpression of RBP-J repressed the SRG3 expression through the N-box motif. Each construct was separately cotransfected with either RBP-J or the empty vector as indicated into 16610D9 cells. Results are the average of three independent experiments. Error bars represent the standard deviation. (c) NotchIC and Deltex1 also downregulated the SRG3 expression even in the presence of the mutation in the N-box sequence. The reporter construct containing N-box mutation of the SRG3 promoter was cotransfected with indicated expression plasmids. Results from three experiments were averaged. Error bars represent standard deviation
Interestingly, we found that Notch1 still repressed the SRG3 promoter activity even in the presence of the N-box mutation (Figure 2c). This raises a possibility that another downstream pathway of Notch1 signaling may play a critical role in repressing the SRG3 expression. To test this possibility, we examined whether Deltex1, another effector molecule of Notch signaling pathway, represses the SRG3 transcription in the presence of the N-box mutation. Overexpression of Deltex1 indeed downregulated the SRG3 expression in the D2/m745 mutants (Figure 2c). These results suggest that repression of the SRG3 expression may occur through Notch-activated RBP-J-dependent and/or RBP-J-independent Deltex1 pathways. However, results from Figures 1 and 2c strongly suggest that Deltex1 may be the primary downstream effector involved in the Notch1-mediated repression of SRG3.
E2A-binding sites in the SRG3 promoter are important for the Notch1-mediated repression
Next, to identify a critical element(s) in the SRG3 promoter responsible for the Notch responsiveness, base substitutions were introduced into the two E-box elements of E558 and E84 (E558 element corresponds to E480 element reported previously13) (Figure 3a). Various constructs containing mutations in the E-boxes (D2/m558, D2/m84, and D2/dm) were transfected into 16610D9 cells together with a plasmid expressing NotchIC or the vector alone. Either mutation at E84 or E558 resulted in a minor repressive effect on the SRG3 promoter activity by NotchIC (Figure 3b). Double mutation at both E84 and E558 (D2/dm) not only reduced the promoter activity (44% of wild type) but also abolished the repressive effect by NotchIC almost completely (Figure 3b). We did not observe any further repressive effect by NotchIC on the D2/dm promoter even in the presence of the N-box motif, which RBP-J appears to act on. These results suggest that the two E-box sequences are critical for the SRG3 promoter activity and that, at least in the 16610D9 cells, they are primary targets for Notch signaling in regulating the SRG3 expression.
E2A-binding sites in the SRG3 promoter are important for the Notch1-mediated repression. (a) Schematic representation of the wild-type (D2) and mutated SRG3 promoter constructs (D2/m84, D2/m558, and D2/dm). Such mutants (○) were generated by base substitutions as described in Materials and Methods. (b) Notch responsiveness of two E-boxes on the SRG3 promoter was examined using various reporter constructs. 16610D9 cells (1 × 107) were cotransfected with 3 μg of indicated luciferase reporter in the presence or absence of 10 μg NotchIC-expressing plasmids. (c) Transcriptional activation of SRG3 by E2A depended on two functional E-box elements. 16610D9 cells were cotransfected with 3 μg of each luciferase reporter, 1 μg of the β-galactosidase expression construct, and indicated amounts of pCAGGS-E47 expression construct. (d) Increased amounts of E47 restored the repression of the SRG3 promoter activity evoked by NotchIC. 16610D9 cells were transfected with D2 reporter construct along with either the empty vector or E47- and/or NotchIC-expressing plasmids, as indicated. The results are representative of three independent experiments. Error bars represent standard deviation
We have previously reported that E2A/HEB protein complex specifically binds to the putative E-box element (E84) in the SRG3 promoter.17, 18 E2A acts as a positive regulator of the SRG3 transcription and its activity is regulated by TCR signaling through Id3 induction during thymocyte maturation. Since we found two E-box elements on the promoter, E84 and E558, we tested whether E47 activates the SRG3 expression through both E-box elements. Ectopic expression of E47 resulted in increased D2 reporter activity in a dose-dependent manner, and similar results were yielded with single E-box mutants (D2/m558 and D2/m84) (Figure 3c). However, the double mutation in both E-boxes (D2/dm) gave no response to increasing E47 expression (Figure 3c). These results suggest that both E-box sequences are important for the regulation of the SRG3 expression.
Next, we examined whether NotchIC specifically represses the E47-dependent transcriptional activation of the SRG3 promoter. To this end, 16610D9 cells were transfected with D2 luciferase reporter plasmid along with various amounts of E47 and/or NotchIC constructs (Figure 3d). The promoter activity of SRG3 decreased significantly by the addition of NotchIC. However, this repression was recovered by increasing the amount of E47 expression, indicating that NotchIC may repress the SRG3 expression by blocking E47 function on the SRG3 promoter.
NotchIC-activated Deltex1 represses the SRG3 expression
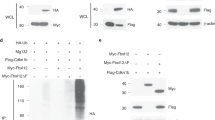
Notch1 was shown to inhibit the E47 activity independently of RBP-J.25 We thus investigated the regulation of the SRG3 expression by Deltex1. By using real-time PCR analysis on 16610D9 cells infected with either retrovirus expressing Deltex1 (S003-Deltex1) or control vector (S003), we found that endogenous SRG3 mRNA was downregulated by Deltex1 (approximately 47% decrease) (Figure 4a). To further confirm that Deltex1 modulates the SRG3 transcription, we have constructed a plasmid expressing a truncated mutant form of Deltex1 (DN-Deltex1), lacking domain III containing a ring-H2-type zinc-finger motif. DN-Deltex1, containing amino acids 1–411 of Deltex1, is known to block homo-oligomerization of Deltex1 and, thus, act as a dominant-negative mutant.21, 29 The repression of the D2 luciferase reporter activity induced by Deltex1 was rescued by increasing amount of DN-Deltex1 (Figure 4b), suggesting that this deletion mutant of Deltex1 has a dominant-negative function on the SRG3 promoter. NotchIC-mediated repression of the SRG3 promoter activity also was rescued by DN-Deltex1 in a dose-dependent manner (Figure 4b). In addition, there was no significant difference between the effects of Deltex1+NotchIC and NotchIC alone on the SRG3 promoter activity (Figure 4b), indicating that Deltex1 did not block the NotchIC inhibitory effect on the SRG3 promoter activity. Although it has previously been reported that Deltex1 partially inhibits Notch1 transactivation,30 Deltex1 did not inhibit Notch-mediated repression of the SRG3 promoter activity in 16610D9 cells. Hence, these results strongly suggest that endogenous Deltex1 induced by NotchIC repressed the SRG3 expression independently of RBP-J. Moreover, the repressive effect of Deltex1 required intact E-box elements in the SRG3 promoter because it was not observed with the double mutant (D2/dm) (Figure 4c). This was already suggested in our previous report30 that both E-box sequences are critical in the control of the SRG3 expression. The SRG3 promoter with mutations at both E84 and E558 did not show any inhibitory response to the Notch1 or Deltex1 treatment. These results suggest that the two E-boxes are major targets of Notch1- and Deltex1-mediated suppression of the SRG3 expression. This is further supported by the results shown in Figure 3c that mutations of two E-boxes gave no response to the increased E47 expression, even though Notch (or Deltex1)-induced repression of SRG3 promoter activity was reversed by increasing the E47 expression (Figures 3d and 4d). From these results, we conclude that the Deltex1 response elements coincide with the two E-box elements (E84 and E558) and they are critical for the Notch responsiveness of the SRG3 promoter. We also observed that ectopic expression of E47 protein blocked the repression by Deltex1 through the two E-box elements, indicating that Deltex1 directly inhibits the E47 function on the SRG3 promoter (Figure 4d).
NotchIC-activated Deltex1 represses the SRG3 expression. (a) 16610D9 cells expressing Deltex1 downregulate SRG3. 16610D9 cells were infected with Flag-Deltex1-IRES-EGFP or control vector-IRES-EGFP. The RNA isolated from these cells was reverse-transcribed and expression of SRG3 was analyzed by real-time PCR analysis. Results were normalized relative to the amount of GAPDH mRNA and were averaged over three independent real-time PCR reactions. Reverse transcribed (RT)-PCR showing the level of transiently expressed flag-tagged Deltex1 (as indicated with arrowhead). (b) Deltex1- or NotchIC-mediated repression of the SRG3 promoter activity was rescued by expression of DN-Deltex1. (c) Repression of the SRG3-luciferase reporter activity mediated by Deltex1 was dependent on the E2A-binding sites (E558, E84). Mutations were introduced into both E-boxes as indicated. (d) Increased amounts of E47 reversed the inhibitory effect of Deltex1 on the SRG3 reporter. 16610D9 cells were transfected with 3 μg of D2 reporter plasmids along with either the empty vector or Deltex1-and/or E47-expressing vectors, as indicated. Luciferase activity was normalized to β-galactosidase activity
Deltex1 downregulates p300-mediated transcriptional activation of the SRG3 promoter
It has been reported that the DNA-binding activity of E2A/HEB heterodimer is modulated upon pre-TCR- and TCR-mediated signaling.31, 32 We have recently shown that the SRG3 transcription is repressed upon TCR activation and Id3 protein induced by TCR activation represses SRG3 transcription by inhibiting the E2A/HEB binding to its promoter.18 In addition, since NotchIC and Deltex1 repress the SRG3 expression through the two E-box sequences on the promoter, we examined how Notch signaling inhibits the transcriptional activity at the molecular level. We first tested whether the Notch signaling inhibits DNA binding of E2A protein to E-box elements on the SRG3 promoter. For this, nuclear extracts of thymocytes from NotchIC transgenic and littermate control mice were incubated with [γ-32P]ATP-radiolabeled E558 and E84 probes, and subjected to electrophoretic mobility shift assay (EMSA) as described in Materials and Methods. Specific DNA–protein complexes for the E558 and E84 probes, reported previously (data not shown),18 were formed with the thymocyte extracts. However, no significant difference in the formation of the complexes was observed between thymocyte extracts from the NotchIC transgenic and control mice (Figure 5a). These results suggest that the E47-binding activity to the promoter is not significantly affected by the Notch signaling.
Deltex1 downregulates p300-mediated transcriptional activation of the SRG3 promoter. (a) EMSA of thymocyte nuclear extracts (5 μg) prepared from C57BL/6 mice (lane 1, 3, and 5) and NotchIC transgenic mice (lane 2, 4, and 6) binding to the E558 and the E84 oligonucleotide probe (left panel). Arrowheads indicate specific complexes. The same extracts used in the left panel were tested for binding to an Oct-1 probe as a control (right panel, lane 5, 6). (b) NotchIC and Deltex1 repressed the interaction of p300 to the E-box element on the SRG3 promoter. In vivo binding of E47 and coactivator p300 to the SRG3 promoter was assayed by ChiP assays. 16610D9 cells were transfected with pCAGGS-Deltex1 and NotchIC, with or without D2 reporter plasmid. Nuclear lysates were incubated with either anti-E47 (3 μg) or anti-p300 (6 μg) antibodies and the co-precipitated SRG3 promoter DNA was amplified using the primers (RV primer3, GL primer2) specific for the promoter region of SRG3 within the plasmid (as indicated with arrowhead). The relative intensity of p300 PCR band was depicted ‘1’ as control. (c) Chromatin immunoprecipitates using the indicated antibodies were probed with primers flanking putative E2A-binding sites in the SRG3 promoter. Thymocytes were derived either from NotchIC transgenic (N) or wild-type mice (C57BL/6). No-Ab group represents a negative control without antibody treatment. (d) ChiP assays were performed on 16610D9 cells infected with a control-(vector) or Deltex-1-expressing retrovirus. Chromatin immunoprecipitates with anti-p300 (2 μg) or without antibody (no α) were probed with primers flanking putative E2A-binding sites on an endogenous SRG3 promoter. Each immunoprecipitated DNA was diluted four-fold, followed by 35 cycles of PCR. As a control, input chromatin that corresponds to 1% of total chromatin was used for each immunoprecipitation. (e) Modulation of E47-dependent transactivation by Deltex1. 293T cells (5 × 105) were transfected with indicated amounts of Deltex1 and p300 along with SRG3 luciferase reporter plasmid. Luciferase activities in whole-cell extracts prepared after 36 h of transfection were measured and normalized to β-galactosidase activity. The data represent results from three independent experiments
Recent reports have also shown that coactivator p300, CBP, and PCAF enhance the E2A-mediated transcriptional activity, and promote nuclear retention of E2A proteins in 293T cells.26 Moreover, p300 can associate with the bHLH region and the amino-terminal region of E2A proteins.21, 25 Furthermore, Deltex1 was reported to bind directly to p300, thereby counteracting p300-dependent transcriptional activation by bHLH protein.21 Therefore, we next examined whether both NotchIC and Deltex1 signaling sequesters the E47 away from coactivators such as p300 stimulating E2A transcriptional activation on the SRG3 promoter. For this, we performed the chromatin immunoprecipitation (ChiP)–PCR assay. As expected, endogenous E47 bound to putative E2A-binding sites in the SRG3 promoter (Figure 5b). Similarly, coactivator p300 was also found to be associated with the SRG3 promoter, presumably via E2A/HEB. However, this association was significantly decreased (62–79%) in the presence of Deltex1 or NotchIC. To further confirm the interaction between Deltex1 and p300 in vivo, we performed ChiP assays using thymocyte extracts from NotchlC transgenic and littermate control mice. Essentially, transgenic NotchIC expression did not affect the DNA binding of E2A protein to the E-box sequences on the endogenous promoter; however, the recruitment of p300 to the protein was significantly blocked (Figure 5c). In addition, the same results were also obtained with Deltex1-transduced 16610D9 cells (Figure 5d). These results indicate that Deltex1 inhibits p300 association on the endogenous SRG3 promoter. We also coexpressed p300 tagged with a HA epitope at its N-terminal with an Myc epitope-tagged Deltex1 in COS-1 cells and confirmed the binding of these proteins by coimmunoprecipitation (data not shown). To support this interaction between Deltex1 and p300, we ectopically expressed either p300 or Deltex1 along with the D2 promoter in 293T cells. While p300 enhanced the SRG3 reporter gene activation by E47 protein in a dose-dependent manner, coexpression of Deltex1 decreased the reporter gene activity (Figure 5e). All these results suggest that the E2A-dependent transcriptional activation of the SRG3 promoter may be regulated by NotchIC-activated Deltex1 by blocking the p300 recruitment.
Enforced expression of Deltex1 restores DP thymocyte survival from the GC-induced apoptosis
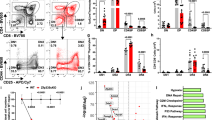
Thymocytes from NotchIC transgenic mice show an increased resistance to the GC-induced apoptosis compared to thymocytes from control mice.10 DP thymocytes from double-transgenic mice overexpressing both SRG3 and NotchIC in T cells became more sensitive to GCs than those from NotchIC transgenic mice.13 Our results suggest that Deltex1 signaling may play an active role in the rescue of DP thymocytes from the GC-induced apoptosis during differentiation of DP thymocytes to SP. To confirm this, flag-tagged Deltex1 was cloned into S003 retroviral vector and transferred into thymocytes from 4- to 7-week-old C57BL/6 mice. Infected thymocytes were stained with Annexin V-PE antibodies and analyzed by flow cytometry after dexamethasone treatment for 3 h. As expected, GC-induced apoptosis of Flag-Deltex1-transduced cells (Deltex1GFP+) was significantly reduced (approximately 71–85% inhibition) as compared with that of control cells (S003GFP−, S003GFP+, and Deltex1GFP−) (Figure 6b). These results suggest that the reduced level of the SRG3 expression by ectopic expression of Deltex1 actually inhibited GC-induced apoptosis in developing thymocytes. It is known that a high percentage of both infected and uninfected thymocytes underwent apoptosis when stimulated with ConA. However, no significant difference was observed in the percentage of apoptotic cells between the infected and uninfected cells.33 Similar results were also obtained with 16610D9 cells which do not require ConA treatment (Figure 6c). However, activation of Notch signal upregulated Bcl-2 in T cells,10 and overexpression of Bcl-2 and Bcl-XL in T cells inhibited GC-induced apoptosis.34, 35 Therefore, it is also possible that GC resistance depends on the level of Bcl-2, not on the level of SRG3. To investigate this possibility, we performed Western blot analysis to see whether expression of NotchIC or Deltex1 affects the expression of Bcl-2 or Bcl-XL. No significant change was observed in the expression of Bcl-2 and Bcl-XL in Deltex1-transduced 16610D9 cells (Figure 6d). However, Bcl-2 was upregulated approximately 2–3-fold in NotchIC transgenic thymocytes (Figure 6e). Therefore, there appeared to be a difference in the expression of Bcl-2 between the transgenic mice and Deltex1-transduced 16610D9 cells. In both systems, cells became resistant to GCs and the expression of SRG3 was downregulated. Detfos et al.10 also reported that cells became resistant to GCs without upregulation of Bcl-2. In addition, we have previously shown that overexpression of SRG3 confers thymocytes a GC resistance without changing the Bcl-2 level.10 These results suggest that thymocytes become resistant to the GC-induced apoptosis by downregulating the SRG3 expression and upregulation of Bcl-2 may not be essential.
Enforced expression of Deltex1 restores DP thymocyte survival from GC-induced apoptosis. (a) Diagram of the retroviral constructs used for the transduction into the thymocytes from C57BL/6 mice and 16610D9 cells. (b, c) Overexpression of Deltex1 in thymocytes from C57BL/6 mice (b) or 16610D9 cells (c) blocks GC-induced apoptosis. Infected cells were stained with Annexin V-PE and analyzed by flow cytometry. Specific apoptosis was calculated as explained in Materials and Methods (◊: S003GFP−, □: S003GFP+, ♦: Deltex1GFP−, ▪: Deltex1GFP+). (d) 16610D9 cells were infected with a control S003 (vector) or S003-Deltex1, and total lysates from these cells were subjected to Western blotting using anti-SRG3, anti-Bcl-XL, anti-Bcl-2, and β-actin antibodies. A representative gel out of three experiments is shown. (e) Thymocyte lysates derived either from NotchIC transgenic or B6 mice (C57BL/6) were immunoblotted with the same antibodies
Discussion
One major phenotypic change that occurs during the differentiation of DP thymocytes into the CD4 and CD8 SP cells is the acquisition of resistance to GC-induced apoptosis. As a result, SP thymocytes become relatively resistant to the GC-induced apoptosis, while DP thymocytes are sensitive to it.11 We have previously shown that SRG3 plays a critical role in determining the GC sensitivity in thymocytes; its expression was downregulated in mature CD4 and CD8 cells,13 and acts as a downstream target of TCR signaling in blocking the GC-induced apoptosis.17, 18 Here, we show that Notch signaling confers developing thymocytes GC resistance also by regulating the SRG3 expression through Deltex1 blocking the recruitment of p300 coactivator to E2A/HEB.
It was suggested that Notch blocks GC-induced apoptosis through the RBP-J pathway of Notch signaling.10 We have identified one N-box sequence in the SRG3 promoter region by homology search, and shown that RBP-J repressed the SRG3 promoter activity through this motif (Figure 2). However, the expression of either Hes1 or Hes5, the downstream effector molecules of RBP-J, did not show any effect on the SRG3 promoter activity in 16610D9 cell line (data not shown). These results suggest the possibility that another gene(s) might be involved in the RBP-J signaling for the repression of the SRG3 transcription.
Even though Deltex1 is positively regulated by Notch signaling in T-cells,10, 20 its role during T-cell development is not clear yet. Deltex1 is expressed at relatively high levels in DN, CD4, and CD8 SP thymocytes, whereas at a low level in DP thymocytes.10, 20 This pattern of Deltex expression suggests that the Notch signaling pathway may operate during differentiation of DN and maturation into CD4/CD8 SP thymocytes. Mice that received Deltex-transduced hematopoietic stem cells (HSC) resulted in a significant increase in the percentage of DN thymocytes.36 Even though overexpression of Deltex1 did not have any functional consequences at DN1–DN3 stages of thymocyte development, percentages of Deltex1-overexpressing DN thymocyte subset population showed an increase in DN3 (53.5 versus 38%) and decrease in DN4 (33 versus 51%) compared to control, suggesting a delay in transition from DN3 to DN4 stages.36 These results suggest a possibility that Deltex1 may actually be involved in the differentiation of thymocytes from DN3 to DN4 stages.
Our observations described here have strong implications for the role of Deltex1 as a regulator of GC sensitivity during thymocyte development. NotchIC and Deltex1 repressed the SRG3 D2/m745 promoter construct efficiently, in which the N-box sequence is disrupted and RBP-J is not functioning. In addition, transfection of NotchIC/ANK, lacking RAM domain, showed an inhibitory effect on the SRG3 promoter activity at a level comparable to NotchIC, whereas the expression of NotchIC/dANK, lacking ankyrin repeats showed only a minor inhibitory effect (Figure 1). We did not observe any significant repressive effect by NotchIC on the D2/dm promoter even though the N-box motif was intact in 16610D9 cells (Figure 3). However, enforced expression of Deltex1 repressed the SRG3 expression in a dose-dependent manner and DN-Deltex1 recovered an inhibitory effect of Notch on the SRG3 promoter (Figure 4). Therefore, it appears that the inhibitory effects of NotchIC on the SRG3 promoter are more dependent on Deltex1 than on RBP-J. It is possible that Deltex1 is preferred in binding to NotchIC to RBP-J, and the repressive effect through RBP-J may be minimal in the presence of Deltex1 at certain stages of differentiation and types of cells.21, 29 It has actually been reported that the expression of an excess amount of Deltex1 in COS-7 cells inhibited co-precipitation of RBP-J with NotchIC.21 In addition, Deltex1 signaling is known to partially inhibit the Notch-mediated RBP-J signaling.29, 30 Furthermore, in support of this notion, we confirmed by real-time PCR analysis that endogenous RBP-J level in NotchIC transgenic mice did not significantly change as compared to that in control mice, but endogenous Deltex1 level increased significantly (Supplementary Figure 1).22 Recently, it was also shown that RBP-J-mediated Notch signaling is dispensable for the lineage commitment, survival, and maturation of CD4/CD8 T-cells.23 In addition, retroviral expression of Deltex1 alone in primary thymocytes and 16610D9 cells was sufficient for the acquisition of resistance to GCs (Figure 6b and c). Therefore, downregulation of the SRG3 expression after positive selection in normal thymocytes is likely to be correlated with the Notch signaling through the Deltex1 activity as well as the TCR signaling.
We have previously shown that E2A/HEB and Id3, induced by TCR signaling and required for proper selections, control the sensitivity to the GC-induced apoptosis in thymocytes by regulating the SRG3 expression.13, 17, 18 In this study, we have shown that Notch signaling directly repressed the E47-dependent SRG3 transcription (Figure 3). These results are consistent with previous reports that Notch and Deltex proteins abrogate E2A-induced transcriptional activation.24, 25, 37 Endogenous E47 bound to two putative E2A-binding sites (E558, E84) in the SRG3 promoter (Figures 5b and c). However, EMSA analysis showed that the E47-binding activity did not change significantly in the presence of the Notch signaling (Figure 5a). Instead, Notch1 and Deltex1 signals might prevent the recruitment of coactivators to E2A, as previously reported that direct interaction between Deltex1 and p300 inhibits transcriptional activation of bHLH proteins.21 We have found that Notch-induced Deltex1 led to direct dissociation of E47 and p300 as a consequence of competitive binding to p300 (Figures 5). Our results, therefore, indicate that the activation of Deltex1 by Notch signaling represses the E47 activity by blocking the recruitment of coactivator p300 to the endogenous SRG3 promoter. On the other hand, in another experimental system, it has been shown that Deltex1 potentiates E2A activity and antagonizes Notch1 signaling.30 Deltex1 partially inhibits the transactivation of an RBP-J-dependent luciferase reporter by NotchIC in several human and murine cells, including B (BJAP) and T (Jurkat) cells. However, a recent report38 showed that the numbers of ectopic B cells in the thymus remained unchanged and B-cell development in the spleen was unaltered in Deltex1 mutant mice. Hence, conclusions of in vivo organ culture and biochemical studies by Izon et al.30 are still controversial. Therefore, the effects of Deltex1 on the transcription activity of E2A proteins appear to vary depending on cell type and developmental stages of cells.
Bcl-2 is expressed throughout T-cell development in both the CD4 and CD8 lineage cells and extends thymocyte survival in a TCR-independent way.34, 35, 39 Both DN and CD4/CD8 SP thymocytes are resistant to GCs and express high levels of Bcl-2 and Deltex1, whereas GC-sensitive DP thymocytes express low levels of Bcl-2 and Deltex1.10 It is possible that Bcl-2 is also involved in the Deltex1-mediated acquisition of GC resistance in thymocytes. No change in the level of Bcl-2 was observed in 16610D9 cells overexpressing Deltex1 (Figure 6d and Supplementary Figure 2). In contrast, Bcl-2 was increased over two-fold in NotchIC transgenic mice (Figure 6e). These data suggest that Deltex1 signaling may confer DP thymocytes a resistance to GCs without upregulating the Bcl-2 and Bcl-XL expression. This is also consistent with the previous report that NotchIC conferred resistance to GCs in 2B4.11 cells in a Bcl-2-independent manner.10 These results suggest that Notch1 regulates the GC sensitivity of thymocytes by repressing the SRG3 expression via Deltex1 and upregulation of Bcl-2 may not be essential for gaining GC resistance.
It is thought that Notch1 may play a role in promoting the maturation of DP thymocytes into both the CD4 and CD8 SP lineages. The expression of Deltex1, activated by Notch1 signaling, is upregulated during the maturation into both CD4 and CD8 SP thymocytes. This suggests a possible role for Deltex1 in promoting the maturation of DP thymocytes into both SP lineages. Our results showed that Notch1 signaling conferred DP thymocyte resistance to GC-induced apoptosis by regulating the SRG3 expression through the Deltex1 signaling pathway. The SRG3 gene is finely regulated by Notch1–Deltex1 signaling as well as the TCR signaling during the maturation from DP to SP thymocytes. Our results identify the SRG3 gene as a target of the Deltex1 and reveal a new function of Deltex1 during thymocyte maturation, inhibiting the transcription activity of E47 by sequestering a coactivator p300. Besides conferring GC resistance, the Notch1–Deltex1 pathway may also contribute to other aspects of the maturation of DP thymocytes into CD4 and CD8 SP cells through the regulation of the SRG3 expression. We recently found that SRG3 overexpression perturbed normal positive and negative selections of DP thymocytes in TCR transgenic mice models (Choi and Seong, in preparation). These results indicate that downregulation of the SRG3 expression is important for proper maturation of DP thymocytes. The SRG3 downexpression occurs right after the selections, probably by both TCR and Notch1 signaling. Therefore, simple overexpression of Deltex1 may not be sufficient to look into its function during the maturation of DP thymocytes. The dual regulatory pathways for the SRG3 expression may also render it difficult to notice the role of the Notch1–Deltex1 signaling in the presence of TCR signaling during the maturation process, unless this targets the expression of gene(s) other than SRG3.
Materials and Methods
Cell lines, transient transfection, and luciferase assay
The murine DP thymoma, 16610D9, was provided by Dr. C Murre (University of California at San Diego, La Jolla, CA, USA) and cultured in Opti-MEM (Invitrogen) containing 10% FBS supplemented with 50 μM 2-mercaptoethanol. 293T cells were maintained in DMEM containing 10% FBS. All media were supplemented with 10 mg/ml penicillin and streptomycin. Transient transfection and reporter assays were performed as described previously.13
DNA constructs
The reporter constructs of the SRG3 promoter were described previously.17 Plasmids encoding human RBP-J were generously provided by T Honjo (Koyto University, Japan). HA-tagged p300 was generously provided by Dr. Y Shi (Harvard Medical School, USA). Mutagenesis of the E-box sequences (CATCTG into CTGCAG) in the SRG3 promoter was performed as described previously. pGL3-Basic vector described in this paper was purchased from Promega. pHBAneo-E47 was obtained from Dr. C Murre. The mouse Notch1 deletion mutants were generated by PCR with the following primers:
- NotchIC/ANK::
-
5′-GATGCTGACTGCATGGATGTCAATGTT-3′, 5′-GAATGCAATTGTTGTTGTTAAC-3′
- NotchIC/ANK2::
-
5′-CCACCATGGATGCCAACATCCAGGAC-3′
- NotchIC/dANK::
-
5′-AACCGGGACATCACGGATCACATG-3′ 5′-GACATCCATGCAGTCAGCATC-3′
The PCR products of both NotchIC/ANK and NotchIC/ANK2 were cloned into pGEM®-T Easy vector (Promega) and then the EcoRI fragment was inserted into pCAGGS vector (generous gift from Dr. Kageyama, Koyto, Japan). The PCR product of NotchIC/dANK was self-ligased and then BamHI fragment was cloned into pCAGGS vector. The mouse Deltex1 cDNA was amplified by RT-PCR and cloned into pGEM®-T Easy vector (Promega), and then the EcoRI fragment was subcloned into pCAGGS vector. S003 is an RZRS-based retrovirus vector with an internal ribosomal entry site (IRES) and GFP as a selection marker in the downstream of the IRES sequence.40 S003-Deltex1 encodes mouse Deltex1 tagged with a Flag epitope. The PCR primers for construction of pCAGGS-Deltex1 and S003-Deltex1 were 5′-CCGGAATTCATGGACTACAAGGACGACGATGACAAGAGATCTATGTCACGGCCAGGCCAAGGT-3′ and 5′-AAGGGAATTCGGGCAACTCAGGCCTCAG-3′. The PCR primers for construction of DN-Deltex1 were 5′-AAAGAATTCCAGGCGGCAGCGGCCATGTC-3′ and 5′-GGTACAGTCCTCATCAGGCGG-3′. The PCR products were verified by sequencing.
Electrophoretic mobility shift assay
Preparation of nuclear extracts was performed as described previously. EMSA was performed as described previously.17, 18 The sequence of each oligonucleotide used for EMSA is as follows:
- E558::
-
5′-CTAAGCAGAAACCATCTGATGTGGTTCGAG-3′
- E84::
-
5′-AGGAGGTGGCATCTGCGCGCGCGG-3′
- Oct-1::
-
5′-TGTCGAATGCAAATCACTAGAA-3′
RNA isolation, RT-PCR, and real-time PCR analysis
Total RNA was prepared from cells using the TRIzol® reagent (Invitrogen) according to the manufacturer's instruction. RT-PCR analysis was performed as described previously. Primers for Bcl-2 were 5′-GGG AGG ATT GTG GCC TTC TT-3′ and 5′-GAT GCC GGT TCA GGT ACT CAG T-3′. Primers for Deltex1 were 5′-CAC TGG CCC TGT CCA CCC AG-3′ and 5′-TAG CTG GCG TCG GGG TAG CC-3′. Primers for HPRT were 5′-GAT ACA GGC CAG ACT TTG TTG-3′ and 5′-GGT AGG CTG GCC TAT AGG CT-3′.
Real-time PCR was performed with a Roto-Gene 3000 (Corbett Research) detection system and SYBR® Green (Qiagen) reagents. The relative gene expression of SRG3 was analyzed using the 2−ΔΔCt method41 by normalizing with GAPDH gene expression in all the experiments. The specific primers were designed for murine SRG3, RBP-J, Deltex1, and GAPDH using Primer3 software program. The following primers were used in real-time PCR:
- SRG3::
-
5′-CTGCATCTTCGATCACCTCA-3′, 5′-CTCCCTTCCAGCACTAGCAC-3′
- Deltex1::
-
5′-CTGGTCACAGCATCTGGCTA-3′, 5′-TGTCTTCTCCCCGTAGATGG-3′
- RBP-J::
-
5′-TCCACCAGCCTTACCTTCAC-3′, 5′-AGTTAGGACACCACGGTTGC-3′
- GAPDH::
-
5′-GAAGGTCGGTGTGAACGAAT-3′, 5′-TTTGATGTTAGCGGGGTCTC-3′
Western blot analysis
Following transduction, 16610D9 cells were lysed as described previously. The lysates (120 μg of protein) were resolved on 12% SDS-PAGE and subjected to standard immunoblotting procedures. The primary antibodies used were anti-SRG3,16 anti-Bcl-XL (Santa Cruz), anti-Bcl-2 (Santa Cruz), and β-actin (Santa Cruz). Protein signals were visualized using the ECL-system (Amersham Pharmacia Biotech).
ChiP assays
ChiP assays were carried out as described previously.16, 42, 43 16610D9 thymoma cells were transfected with or without 10 μg pGL-D2 (−1145 to −1). After incubation at 37°C for 24 h, cells were treated with 1% formaldehyde for 15 min at 37°C. Then, cells were collected and resuspended in lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl (pH 8.1)) with 1 mM PMSF, pepstatin A, and aprotinin. After brief sonication, lysates were cleared by centrifugation. The lysates were diluted 10-fold with dilution buffer (0.01% SDS, 1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8.1, and 150 mM NaCl). The lysates were incubated with either anti-E47 (Pharmingen) or anti-p300 (Santa Cruz) antibodies with rotation at 4°C overnight. Immunoprecipitated complexes were collected by protein A sepharose beads. Immunoprecipitates were washed three times with wash buffer and twice with TE buffer. Immunocomplexes were eluted with 2 volume of 250 μl of elution buffer (1% SDS, 0.1 M NaHCO3) at room temperature for 15 min with rotation. Then, 20 μl of 5 M NaCl was added to reverse formaldehyde crosslinking by heating at 65°C for 4 h. Pellets were resuspended and treated with proteinase K for 1 h at 45°C. DNA was extracted with phenol/chloroform and precipitated with ethanol. Pellets were resuspended in 10 mM Tris-HCl and 1 mM EDTA and amplified by 35 cycles of PCR with primers corresponding to the flanking region of SRG3 promoter in the pGL3 basic vector (Promega).
The primer for RV primer3 was 5′-CTAGCAAAATAGGCTGTCCC-3′ and that for GL primer2 was 5′-CTTTATGTTTTTGGCGTCTTCCA-3′. The E558 primers used in ChiP of mice were 5′-CACCAGGTTGAAGAGAGAGCTA-3′ (−739 to −718) and 5′-CACCCCCGAGCGCTAGTAAG-3′ (−440 to −470). Primers for E84 were 5′-AAAGGCGGGCAGCCTCAGTC-3′ (−249 to −230) and E84-2 5′-ATAGTCACCGTCCGTCGTGTC-3′ (+2 to −19).
Retroviral transduction and cell transduction
The NX-eco packaging cell line was transfected with the retroviral construct by calcium phosphate precipitation as described previously.17 Retrovirus-containing supernatant was collected after overnight incubation of the transfected packaging cells at 32°C. Thymocytes from 4- to 7-week-old C57BL/6 mice at 5 × 106 were infected in retrovirus-containing supernatant supplemented with 8 μg/ml polybrene, followed by centrifugation at 2800 r.p.m. for 90 min at 30–32°C and overnight incubation at 37°C. At 2 days following infection, dexamethasone was added to the cultures in six-well plates at final concentrations of 10−6, 10−7, and 10−8 M for 3 h.
Flow cytometry
Single-cell suspensions of thymocytes derived from 4- to 5-week-old wild-type C57BL/6 mice were maintained in RPMI 1640 medium containing 10% FBS and 50 μM 2-mercaptoethanol supplemented with glutamine, 10 mg/ml penicillin, and streptomycin. The portion of apoptotic cells were assessed by Annexin V staining. At the end of each treatment, cells were resuspended in Annexin V-PE, incubated at room temperature for 15 min, washed, resuspended in 1 × binding buffer (10 mM Hepes/NaOH (pH 7.4), 140 mM NaCl, 2.5 mM CaCl2) and analyzed by flow cytometry (50 000–70 000 events) with the CellQuest™ software using FACStar (Becton Dickinson). Percent specific apoptosis was calculated by the formula16; (Experimental apoptosis−Spontaneous apoptosis)/(100−Spontaneous apoptosis) × 100 (%).
Abbreviations
- TCR:
-
T-cell receptor
- GC:
-
glucocorticoid
- DP:
-
double positive
- DN:
-
double negative
- SP:
-
single positive
- NotchIC:
-
activated form of Notch1
- DN-Deltex1:
-
dominant-negative form of Deltex1
References
Schwartz RH (1985) T-lymphocyte recognition of antigen in association with gene products of the major histocompatibility complex. Annu. Rev. Immunol. 3: 237–261
von Boehmer H (1994) Positive selection of lymphocytes. Cell 76: 219–228
Marrack P and Kappler J (1987) The T cell receptor. Science 238: 1073–1079
Sprent J, Lo D, Gao EK and Ron Y (1988) T cell selection in the thymus. Immunol. Rev. 101: 173–190
Izon DJ, Punt JA, Xu L, Karnell FG, Allman D, Myung PS, Boerth NJ, Pui JC, Koretzky GA and Pear WS (2001) Notch1 regulates maturation of CD4+ and CD8+ thymocytes by modulating TCR signal strength. Immunity 14: 253–264
Radtke F, Wilson A, Mancini SJ and MacDonald HR (2004) Notch regulation of lymphocyte development and function. Nat. Immunol. 5: 247–253
Walunas TL, Sperling AI, Khattri R, Thompson CB and Bluestone JA (1996) CD28 expression is not essential for positive and negative selection of thymocytes or peripheral T cell tolerance. J. Immunol. 156: 1006–1013
Yasutomo K, Doyle C, Miele L, Fuchs C and Germain RN (2000) The duration of antigen receptor signalling determines CD4+ versus CD8+ T-cell lineage fate. (erratum appears in Nature 2000; 407: 110). Nature 404: 506–510
Cohen JJ and Duke RC (1984) Glucocorticoid activation of a calcium-dependent endonuclease in thymocyte nuclei leads to cell death. J. Immunol. 132: 38–42
Deftos ML, He YW, Ojala EW and Bevan MJ (1998) Correlating notch signaling with thymocyte maturation. Immunity 9: 777–786
Vacchio MS and Ashwell JD (2000) Glucocorticoids and thymocyte development. Semin. Immunol. 12: 475–485
Wagner Jr DH, Hagman J, Linsley PS, Hodsdon W, Freed JH and Newell MK (1996) Rescue of thymocytes from glucocorticoid-induced cell death mediated by CD28/CTLA-4 costimulatory interactions with B7-1/B7-2. J. Exp. Med. 184: 1631–1638
Choi YI, Jeon SH, Jang J, Han S, Kim JK, Chung HY, Park SD and Seong RH (2001) Notch1 confers a resistance to glucocorticoid-induced apoptosis on developing thymocytes by down-regulating SRG3 expression. Proc. Natl. Acad. Sci. USA. 98: 10267–10272
Jeon SH, Kang MG, Kim YH, Jin YH, Lee C, Chung HY, Kwon H, Park SD and Seong RH (1997) A new mouse gene, SRG3, related to the SWI3 of Saccharomyces cerevisiae, is required for apoptosis induced by glucocorticoids in a thymoma cell line. J. Exp. Med. 185: 1827–1836
Kim JK, Huh SO, Choi H, Lee KS, Shin D, Lee C, Nam JS, Kim H, Chung H, Lee HW, Park SD and Seong RH (2001) Srg3, a mouse homolog of yeast SWI3, is essential for early embryogenesis and involved in brain development. Mol. Cell. Biol. 21: 7787–7795
Han S, Choi H, Ko MG, Choi YI, Sohn DH, Kim JK, Shin D, Chung H, Lee HW, Kim JB, Park SD and Seong RH (2001) Peripheral T cells become sensitive to glucocorticoid- and stress-induced apoptosis in transgenic mice overexpressing SRG3. J. Immunol. 167: 805–810
Ko M, Jang J, Ahn J, Lee K, Chung H, Jeon SH and Seong RH (2004) T cell receptor signaling inhibits glucocorticoid-induced apoptosis by repressing the SRG3 expression via Ras activation. J. Biol. Chem. 279: 21903–21915
Ko M, Ahn J, Lee C, Chung H, Jeon SH, Chung HY and Seong RH (2004) E2A/HEB and Id3 proteins control the sensitivity to glucocorticoid-induced apoptosis in thymocytes by regulating the SRG3 expression. J. Biol. Chem. 279: 21916–21923
Jeong SM, Lee KY, Shin D, Chung H, Jeon SH and Seong RH (2004) Nitric oxide inhibits glucocorticoid-induced apoptosis of thymocytes by repressing the SRG3 expression. J. Biol. Chem. 279: 34373–34379
Deftos ML, Huang E, Ojala EW, Forbush KA and Bevan MJ (2000) Notch1 signaling promotes the maturation of CD4 and CD8 SP thymocytes. Immunity 13: 73–84
Yamamoto N, Yamamoto S, Inagaki F, Kawaichi M, Fukamizu A, Kishi N, Matsuno K, Nakamura K, Weinmaster G, Okano H and Nakafuku M (2001) Role of Deltex-1 as a transcriptional regulator downstream of the Notch receptor. J. Biol. Chem. 276: 45031–45040
Saito T, Chiba S, Ichikawa M, Kunisato A, Asai T, Shimizu K, Yamaguchi T, Yamamoto G, Seo S, Kumano K, Nakagami-Yamaguchi E, Hamada Y, Aizawa S and Hirai H (2003) Notch2 is preferentially expressed in mature B cells and indispensable for marginal zone B lineage development (see comment). Immunity 18: 675–685
Tanigaki K, Tsuji M, Yamamoto N, Han H, Tsukada J, Inoue H, Kubo M and Honjo T (2004) Regulation of alphabeta/gammadelta T cell lineage commitment and peripheral T cell responses by Notch/RBP-J signaling. Immunity 20: 611–622
Matsuno K, Eastman D, Mitsiades T, Quinn AM, Carcanciu ML, Ordentlich P, Kadesch T and Artavanis-Tsakonas S (1998) Human deltex is a conserved regulator of Notch signalling. Nat. Genet. 19: 74–78
Ordentlich P, Lin A, Shen CP, Blaumueller C, Matsuno K, Artavanis-Tsakonas S and Kadesch T (1998) Notch inhibition of E47 supports the existence of a novel signaling pathway. Mol. Cell. Biol. 18: 2230–2239
Bradney C, Hjelmeland M, Komatsu Y, Yoshida M, Yao TP and Zhuang Y (2003) Regulation of E2A activities by histone acetyltransferases in B lymphocyte development. J. Biol. Chem. 278: 2370–2376
Qiu Y, Guo M, Huang S and Stein R (2002) Insulin gene transcription is mediated by interactions between the p300 coactivator and PDX-1, BETA2, and E47. Mol. Cell. Biol. 22: 412–420
Ohtsuka T, Ishibashi M, Gradwohl G, Nakanishi S, Guillemot F and Kageyama R (1999) Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. EMBO J. 18: 2196–2207
Hu QD, Ang BT, Karsak M, Hu WP, Cui XY, Duka T, Takeda Y, Chia W, Sankar N, Ng YK, Ling EA, Maciag T, Small D, Trifonova R, Kopan R, Okano H, Nakafuku M, Chiba S, Hirai H, Aster JC, Schachner M, Pallen CJ, Watanabe K and Xiao ZC (2003) F3/contactin acts as a functional ligand for Notch during oligodendrocyte maturation. Cell 115: 163–175
Izon DJ, Aster JC, He Y, Weng A, Karnell FG, Patriub V, Xu L, Bakkour S, Rodriguez C, Allman D and Pear WS (2002) Deltex1 redirects lymphoid progenitors to the B cell lineage by antagonizing Notch1. Immunity 16: 231–243
Bain G, Cravatt CB, Loomans C, Alberola-Ila J, Hedrick SM and Murre C (2001) Regulation of the helix–loop–helix proteins, E2A and Id3, by the Ras-ERK MAPK cascade. Nat. Immunol. 2: 165–171
Engel I, Johns C, Bain G, Rivera RR and Murre C (2001) Early thymocyte development is regulated by modulation of E2A protein activity. J. Exp. Med. 194: 733–745
Joshi A, Vahlenkamp TW, Garg H, Tompkins WA and Tompkins MB (2004) Preferential replication of FIV in activated CD4(+)CD25(+)T cells independent of cellular proliferation. Virology 321: 307–322
Sentman CL, Shutter JR, Hockenbery D, Kanagawa O and Korsmeyer SJ (1991) bcl-2 inhibits multiple forms of apoptosis but not negative selection in thymocytes. Cell 67: 879–888
Schmidt S, Rainer J, Ploner C, Presul E, Riml S and Kofler R (2004) Glucocorticoid-induced apoptosis and glucocorticoid resistance: molecular mechanisms and clinical relevance. Cell Death Differ. 11 (Suppl 1): S45–S55
Yun TJ and Bevan MJ (2003) Notch-regulated ankyrin-repeat protein inhibits Notch1 signaling: multiple Notch1 signaling pathways involved in T cell development. J. Immunol. 170: 5834–5841
Kishi N, Tang Z, Maeda Y, Hirai A, Mo R, Ito M, Suzuki S, Nakao K, Kinoshita T, Kadesch T, Hui C, Artavanis-Tsakonas S, Okano H and Matsuno K (2001) Murine homologs of deltex define a novel gene family involved in vertebrate Notch signaling and neurogenesis. Int. J. Dev. Neurosci. 19: 21–35
Storck S, Delbos F, Stadler N, Thirion-Delalande C, Bernex F, Verthuy C, Ferrier P, Weill JC and Reynaud CA (2005) Normal immune system development in mice lacking the Deltex-1 RING finger domain. Mol. Cell. Biol. 25: 1437–1445
Linette GP, Grusby MJ, Hedrick SM, Hansen TH, Glimcher LH and Korsmeyer SJ (1994) Bcl-2 is upregulated at the CD4+ CD8+ stage during positive selection and promotes thymocyte differentiation at several control points. Immunity 1: 197–205
Bain G, Quong MW, Soloff RS, Hedrick SM and Murre C (1999) Thymocyte maturation is regulated by the activity of the helix–loop–helix protein, E47. J. Exp. Med. 190: 1605–1616
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29: e45
Kim HP, Kelly J and Leonard WJ (2001) The basis for IL-2-induced IL-2 receptor alpha chain gene regulation: importance of two widely separated IL-2 response elements. Immunity 15: 159–172
Youn HD and Liu JO (2000) Cabin1 represses MEF2-dependent Nur77 expression and T cell apoptosis by controlling association of histone deacetylases and acetylases with MEF2. Immunity 13: 85–94
Acknowledgements
This work was supported in part by a grant from the Korea Science and Engineering Foundation, through the Research Center for Functional Cellulomics. JJ, YC, KYL, and SHJ were supported by the BK21, Research Fellowship from Korea Ministry of Education and Human Resources.
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by BA Osborne
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Supplementary information
Rights and permissions
About this article
Cite this article
Jang, J., Choi, Y., Choi, J. et al. Notch1 confers thymocytes a resistance to GC-induced apoptosis through Deltex1 by blocking the recruitment of p300 to the SRG3 promoter. Cell Death Differ 13, 1495–1505 (2006). https://doi.org/10.1038/sj.cdd.4401827
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.cdd.4401827
Keywords
This article is cited by
-
Functional impact of NOTCH1 mutations in chronic lymphocytic leukemia
Leukemia (2014)
-
Luteinizing Hormone Receptor Deficiency Increases the Susceptibility to Alkylating Agent-Induced Lymphomagenesis in Mice
Hormones and Cancer (2010)
-
γ-secretase inhibitors reverse glucocorticoid resistance in T cell acute lymphoblastic leukemia
Nature Medicine (2009)
-
Notch signalling suppresses apoptosis in adult human and mouse pancreatic islet cells
Diabetologia (2007)