Abstract
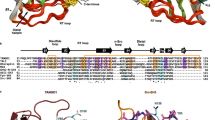
GGAs (Golgi-localizing, γ-adaptin ear homology domain, ARF-interacting proteins) are critical for the transport of soluble proteins from the trans-Golgi network (TGN) to endosomes/lysosomes by means of interactions with TGN-sorting receptors, ADP-ribosylation factor (ARF), and clathrin1,2. The amino-terminal VHS domains of GGAs form complexes with the cytoplasmic domains of sorting receptors by recognizing acidic-cluster dileucine (ACLL) sequences1,2,3,4,5,6. Here we report the X-ray structure of the GGA1 VHS domain alone, and in complex with the carboxy-terminal peptide of cation-independent mannose 6-phosphate receptor containing an ACLL sequence. The VHS domain forms a super helix with eight α-helices, similar to the VHS domains of TOM1 and Hrs. Unidirectional movements of helices α6 and α8, and some of their side chains, create a set of electrostatic and hydrophobic interactions for correct recognition of the ACLL peptide. This recognition mechanism provides the basis for regulation of protein transport from the TGN to endosomes/lysosomes, which is shared by sortilin and low-density lipoprotein receptor-related protein.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Black, M. W. & Pelham, H. R. B. B. Membrane traffic: how do GGAs fit in with the adaptors? Curr. Biol. 11, R460–R462 (2001).
Robinson, M. S. & Bonifacino, J. S. Adaptor-related proteins. Curr. Opin. Cell Biol. 13, 444–453 (2001).
Nielsen, M. S. et al. The sortilin cytoplasmic tail conveys Golgi-endosome transport and binds the VHS domain of the GGA2 sorting protein. EMBO J. 20, 2180–2190 (2001).
Puertollano, R., Aguilar, R. C., Gorshkova, I., Crouch, R. J. & Bonifacino, J. S. Sorting of mannose 6-phosphate receptors mediated by the GGAs. Science 292, 1712–1716 (2001).
Takatsu, H., Katoh, Y., Shiba, Y. & Nakayama, K. Golgi-localizing, γ-adaptin ear homology domain, ADP-ribosylation factor binding (GGA) proteins interact with acidic dileucine sequences within the cytoplasmic domains of sorting receptors through their Vps27p/Hrs/STAM (VHS) domains. J. Biol. Chem. 276, 28541–28545 (2001).
Zhu, Y., Doray, B., Poussu, A., Lehto, V. P. & Kornfeld, S. Binding of GGA2 to the lysosomal enzyme sorting motif of the mannose 6-phosphate receptor. Science 292, 1716–1718 (2001).
Meyer, C. et al. µ1A-adaptin-deficient mice: lethality, loss of AP-1 binding and rerouting of mannose 6-phosphate receptors. EMBO J. 19, 2193–2203 (2000).
Chen, H. J., Yuan, J. & Lobel, P. Systematic mutational analysis of the cation-independent mannose 6-phosphate/insulin-like growth factor II receptor cytoplasmic domain: an acidic cluster containing a key aspartate is important for function in lysosomal enzyme sorting. J. Biol. Chem. 272, 7003–7012 (1997).
Boman, A. L., Zhang, C.-J., Zhu, X. & Kahn, R. A. A family of ADP-ribosylation factor effectors that can alter membrane transport through the trans-Golgi. Mol. Biol. Cell 11, 1241–1255 (2000).
Dell'Angelica, E. C. et al. GGAs: a family of ADP ribosylation factor-binding proteins related to adaptors and associated with the Golgi complex. J. Cell Biol. 149, 81–83 (2000).
Hirst, J. et al. A family of proteins with γ-adaptin and VHS domains that facilitate trafficking between the trans-Golgi network and the vacuole/lysosome. J. Cell Biol. 149, 67–79 (2000).
Poussu, A., Lohi, O. & Lehto, V. -P. Vear, a novel Golgi-associated protein with VHS and γ-adaptin ‘ear’ domains. J. Biol. Chem. 275, 7176–7183 (2000).
Takatsu, H., Yoshino, K. & Nakayama, K. Adaptor-γ ear homology domain conserved in γ-adaptin and GGA proteins that interact with γ-synergin. Biochem. Biophys. Res. Commun. 271, 719–725 (2000).
Zhdankina, O., Strand, N. L., Redmond, J. M. & Boman, A. L. Yeast GGA proteins interact with GTP-bound Arf and facilitate transport through the Golgi. Yeast 18, 1–18 (2001).
Costaguta, G., Stefan, C. J., Bensen, E. S., Emr, S. D. & Payne, G. S. Yeast Gga coat proteins function with clathrin in Golgi to endosome transport. Mol. Biol. Cell 12, 1885–1896 (2001).
Puertollano, R., Randazzo, P. A., Presley, J. F., Hartnell, L. M. & Bonifacino, J. S. The GGAs promote ARF-dependent recruitment of clathrin to the TGN. Cell 105, 93–102 (2001).
Lohi, O. & Lehto, V.-P. VHS domain marks a group of proteins involved in endocytosis and vesicular trafficking. FEBS Lett. 440, 255–257 (1998).
Mao, Y. et al. Crystal structure of the VHS and FYVE tandem domains of Hrs, a protein involved in membrane trafficking and signal transduction. Cell 100, 447–456 (2000).
Misra, S., Beach, B. M. & Hurley, J. H. Structure of the VHS domain of human Tom1 (Target of Myb1): insights into interactions with proteins and membranes. Biochemistry 39, 11282–11290 (2000).
Otwinoski, Z. in Data Collection and Processing (eds Sawyer, L., Isaacs, N. & Bailey, S.) 56–62 (SERC Daresbury Laboratory, Warrington, 1993).
Leslie, A. W. G. Joint CCP4 and ESF-EACMB Newsletter on Protein Crystallography Vol. 26 (Daresbury Laboratory, Warrington, 1992).
Evans, P. R. Proceedings of the CCP4 Study Weekend on Data Collection & Processing 114–122 (Daresbury Laboratory, Warrington, 1993).
Brünger, A. T. et al. Crystallography & NMR system: a new software for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998).
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997).
Navaza, J. AMORE—an automated package for molecular replacement. Acta Crystallogr. A 50, 157–163 (1994).
Roussel, A. & Cambillau, C. Silicon Graphics Geometry Partners (Silicon Graphics, Mountain View, California, 1991).
Kraulis, P. J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946–950 (1991).
Nicholls, A., Sharp, K. A. & Honig, B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins Struct. Funct. Genet. 11, 281–296 (1991).
Wallace, A. C., Laskowski, R. A. & Thornton, J. M. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein. Eng. 8, 127–134 (1995).
Acknowledgements
This work was supported in part by Grants-in Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan, from the Japan Society for Promotion of Science (fellowship to H.T.), and from the University of Tsukuba Research Projects.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Shiba, T., Takatsu, H., Nogi, T. et al. Structural basis for recognition of acidic-cluster dileucine sequence by GGA1. Nature 415, 937–941 (2002). https://doi.org/10.1038/415937a
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/415937a
This article is cited by
-
A Naturally Occurring Splice Variant of GGA1 Inhibits the Anterograde Post-Golgi Traffic of α2B-Adrenergic Receptor
Scientific Reports (2019)
-
Preferential phosphatidylinositol 5-phosphate binding contributes to a destabilization of the VHS domain structure of Tom1
Scientific Reports (2019)
-
Regulation of α2B-Adrenergic Receptor Cell Surface Transport by GGA1 and GGA2
Scientific Reports (2016)
-
Evolutionary analysis of the ENTH/ANTH/VHS protein superfamily reveals a coevolution between membrane trafficking and metabolism
BMC Genomics (2012)
-
Structural basis of cargo recognition by the myosin-X MyTH4-FERM domain
The EMBO Journal (2011)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.