Abstract
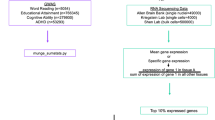
Quantitative genetic research suggests that reading disability is the quantitative extreme of the same genetic and environmental factors responsible for normal variation in reading ability. This finding warrants a quantitative trait locus (QTL) strategy that compares low versus high extremes of the normal distribution of reading in the search for QTLs associated with variation throughout the distribution. A low reading ability group (N=755) and a high reading group (N=747) were selected from a representative UK sample of 7-year-olds assessed on two measures of reading that we have shown to be highly heritable and highly genetically correlated. The low and high reading ability groups were each divided into 10 independent DNA pools and the 20 pools were assayed on 100 K single nucleotide polymorphism (SNP) microarrays to screen for the largest allele frequency differences between the low and high reading ability groups. Seventy five of these nominated SNPs were individually genotyped in an independent sample of low (N=452) and high (N=452) reading ability children selected from a second sample of 4258 7-year-olds. Nine of the seventy-five SNPs were nominally significant (P<0.05) in the predicted direction. These 9 SNPs and 14 other SNPs showing low versus high allele frequency differences in the predicted direction were genotyped in the rest of the second sample to test the QTL hypothesis. Ten SNPs yielded nominally significant linear associations in the expected direction across the distribution of reading ability. However, none of these SNP associations accounted for more than 0.5% of the variance of reading ability, despite 99% power to detect them. We conclude that QTL effect sizes, even for highly heritable common disorders and quantitative traits such as early reading disability and ability, might be much smaller than previously considered.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Shaywitz SE . Dyslexia. New Engl J Med 1998; 338: 307–312.
Snowling M . Dyslexia, 2nd edn. Blackwell: Oxford, 2000.
Jacobson C . How persistent is reading disability? Individual growth curves in reading. Dyslexia 1999; 5: 78–93.
Shaywitz SE, Fletcher JM, Holahan JM, Shneider AE, Marchione KE, Stuebing KK et al. Persistence of dyslexia: the Connecticut Longitudinal Study at adolescence. Pediatrics 1999; 104: 1351–1359.
DeFries JC, Knopik VS, Wadsworth SJ . Colorado Twin Study of reading disability. In: Duane DD (ed). Reading and Attention Disorders: Neurobiological Correlates. York Press: Baltimore Maryland, 1999, pp 17–41.
Harlaar N, Spinath FM, Dale PS, Plomin R . Genetic influences on early word recognition abilities and disabilities: a study of 7-year-old twins. J Child Psychol Psychiatry 2005; 46: 373–384.
Plomin R, Kovas Y . Generalist genes and learning disabilities. Psychol Bull 2005; 131: 592–617.
Plomin R, Owen MJ, McGuffin P . The genetic basis of complex human behaviors. Science 1994; 264: 1733–1739.
Paracchini S, Scerri T, Monaco AP . The genetic lexicon of dyslexia. Annu Rev Genomics Hum Genet 2007 [E-pub ahead of print].
Fisher SE, Francks C . Genes, cognition and dyslexia: learning to read the genome. Trends Cogn Sci 2006; 10: 250–257.
Williams J, O'Donovan MC . The genetics of developmental dyslexia. Eur J Hum Genet 2006; 14: 681–689.
McGrath LM, Smith SD, Pennington BF . Breakthroughs in the search for dyslexia candidate genes. Trends Mol Med 2006; 12: 333–341.
Schumacher J, Hoffmann P, Schmael C, Schulte-Korne G, Nothen MM . Genetics of dyslexia: the evolving landscape. J Med Genet 2007; 44: 289–297.
Deffenbacher KE, Kenyon JB, Hoover DM, Olson RK, Pennington BF, DeFries JC et al. Refinement of the 6p21.3 quantitative trait locus influencing dyslexia: linkage and association analyses. Hum Genet 2004; 115: 128–138.
Risch NJ . Searching for genetic determinants in the new millennium. Nature 2000; 405: 847–856.
Sham PC, Cherny SS, Purcell S, Hewitt JK . Power of linkage versus association analysis of quantitative traits, by use of variance-components models, for sibship data. Am J Hum Genet 2000; 66: 1616–1630.
Zondervan KT, Cardon LR . The complex interplay among factors that influence allelic association. Nat Rev Genet 2004; 5: 89–100.
Cardon LR, Bell JI . Association study designs for complex diseases. Nat Rev Genet 2001; 2: 91–99.
Ioannidis JP, Trikalinos TA, Khoury MJ . Implications of small effect sizes of individual genetic variants on the design and interpretation of genetic association studies of complex diseases. Am J Epidemiol 2006; 164: 609–614.
Syvanen AC . Toward genome-wide SNP genotyping. Nat Genet 2005; 37 (Suppl): S5–S10.
Hirschhorn JN, Daly MJ . Genome-wide association studies for common diseases and complex traits. Nat Rev Genet 2005; 6: 95–108.
Carlson CS, Eberle MA, Kruglyak L, Nickerson DA . Mapping complex disease loci in whole-genome association studies. Nature 2004; 429: 446–452.
Newton-Cheh C, Hirschhorn JN . Genetic association studies of complex traits: design and analysis issues. Mutat Res 2005; 573: 54–69.
Thomas DC, Haile RW, Duggan D . Recent developments in genomewide association scans: a workshop summary and review. Am J Hum Genet 2005; 77: 337–345.
Wang WY, Barratt BJ, Clayton DG, Todd JA . Genome-wide association studies: theoretical and practical concerns. Nat Rev Genet 2005; 6: 109–118.
Knight J, Sham P . Design and analysis of association studies using pooled DNA from large twin samples. Behav Genet 2006; 36: 665–677.
Darvasi A, Soller M . Selective DNA pooling for determination of linkage between a molecular marker and a quantitative trait locus. Genetics 1994; 138: 1365–1373.
Norton N, Williams NM, O'Donovan MC, Owen MJ . DNA pooling as a tool for large-scale association studies in complex traits. Ann Med 2004; 36: 146–152.
Butcher LM, Meaburn E, Liu L, Fernandes C, Hill L, Al Chalabi A et al. Genotyping pooled DNA on microarrays: a systematic genome screen of thousands of SNPs in large samples to detect QTLs for complex traits. Behav Genet 2004; 34: 549–555.
Meaburn E, Butcher LM, Liu L, Fernandes C, Hansen V, Al Chalabi A et al. Genotyping DNA pools on microarrays: tackling the QTL problem of large samples and large numbers of SNPs. BMC Genomics 2005; 6: 52.
Meaburn E, Butcher LM, Schalkwyk LC, Plomin R . Genotyping pooled DNA using 100K SNP microarrays: a step towards genomewide association scans. Nucleic Acids Res 2006; 34: e27.
Pearson JV, Huentelman MJ, Halperin RF, Tembe WD, Melquist S, Homer N et al. Identification of the genetic basis for complex disorders by use of pooling-based genomewide single-nucleotide-polymorphism association studies. Am J Hum Genet 2007; 80: 126–139.
Kirov G, Nikolov I, Georgieva L, Moskvina V, Owen MJ, O'Donovan MC . Pooled DNA genotyping on Affymetrix SNP genotyping arrays. BMC Genomics 2006; 7: 27.
Butcher LM, Meaburn E, Knight J, Sham PC, Schalkwyk LC, Craig IW et al. SNPs, microarrays, and pooled DNA: identification of four loci associated with mild mental impairment in a sample of 6000 children. Hum Mol Genet 2005; 14: 1315–1325.
Evans DM, Cardon LR, Morris AP . Genotype prediction using a dense map of SNPs. Genet Epidemiol 2004; 27: 375–384.
Ke X, Hunt S, Tapper W, Lawrence R, Stavrides G, Ghori J et al. The impact of SNP density on fine-scale patterns of linkage disequilibrium. Hum Mol Genet 2004; 13: 577–588.
Oliver B, Plomin R . Twins' Early Development Study (TEDS): a multivariate, longitudinal genetic investigation of language, cognition and behavior problems from childhood through adolescence. Twin Res Hum Genet 2007; 10: 96–105.
Sham P, Bader JS, Craig I, O'Donovan M, Owen M . DNA Pooling: a tool for large-scale association studies. Nat Rev Genet 2002; 3: 862–871.
Torgesen JK, Wagner RK, Rashotte CA . Test of Word Reading Efficiency. Pro-Ed: Austin, TX, 1999.
Qualifications and Curriculum Authority. QCA Key Stage 1: Assessment and reporting arrangements. Great Britain Qualifications and Curriculum Authority 1999.
Dale PS, Harlaar N, Plomin R . Telephone testing and teacher assessment of reading skills in 7-year-olds: I. Substantial correspondence for a sample of 5808 Children And for extremes. Reading and Writing: An Interdisciplinary Journal 2005; 18: 385–400.
Harlaar N, Dale PS, Plomin R . Correspondence between telephone testing and teacher assessments of reading skills in a sample of 7-year-old twins: II. Strong genetic overlap. Reading and writing: An interdisciplinary journal 2005; 18: 401–423.
Lyon JR, Fletcher JM, Shaywitz BA, Shaywitz SE, Torgesen JK, Wood FB et al. Rethinking special education for a new century. In: Finn CE, Rotherham RAJ, Hokanson CR (eds). Rethinking Learning Disabilities, Thomas B. Fordham Foundation and Progressive Policy Institute: Washington, DC, 2001, pp 259–287.
Freeman B, Smith N, Curtis C, Huckett L, Mill J, Craig I . DNA from buccal swabs recruited by mail: evaluation of storage effects on long-term stability and suitability for multiplex polymerase chain reaction genotyping. Behav Genet 2003; 33: 67–72.
Matsuzaki H, Dong S, Loi H, Di X, Liu G, Hubbell E et al. Genotyping over 100 000 SNPs on a pair of oligonucleotide arrays. Nat Methods 2004; 1: 109–111.
Tobler AR, Short S, Andersen MR, Paner TM, Briggs JC, Lambert SM et al. The SNPlex genotyping system: a flexible and scalable platform for SNP genotyping. J Biomol Tech 2005; 16: 398–406.
Cohen J . Statistical Power Analysis for the Behavioral Sciences. Academic Press: New York, 2004.
Lau W, Kuo TY, Tapper W, Cox S, Collins A . Exploiting large scale computing to construct high resolution linkage disequilibrium maps of the human genome. Bioinformatics 2007; 23: 517–519.
Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C et al. Complement factor H polymorphism in age-related macular degeneration. Science 2005; 308: 385–389.
Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science 2006; 314: 1461–1463.
Plomin R, DeFries JC, Craig IW, McGuffin P . Behavioral genetics. In: Plomin R, DeFries JC, Craig IW, McGuffin P (eds). Behavioral Genetics in the Postgenomic Era. American Psychological Association: Washington, DC, 2003, pp 3–15.
Simpson CL, Knight J, Butcher LM, Hansen VK, Meaburn E, Schalkwyk LC et al. A central resource for accurate allele frequency estimation from pooled DNA genotyped on DNA microarrays. Nucleic Acids Res 2005; 33: e25.
Cargill M, Daley GQ . Mining for SNPs: putting the common variants--common disease hypothesis to the test. Pharmacogenomics 2000; 1: 27–37.
Craig DW, Huentelman MJ, Hu-Lince D, Zismann VL, Kruer MC, Lee AM et al. Identification of disease causing loci using an array-based genotyping approach on pooled DNA. BMC Genomics 2005; 6: 138.
Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD et al. Global variation in copy number in the human genome. Nature 2006; 444: 444–454.
Wong KK, deLeeuw RJ, Dosanjh NS, Kimm LR, Cheng Z, Horsman DE et al. A comprehensive analysis of common copy-number variations in the human genome. Am J Hum Genet 2007; 80: 91–104.
Acknowledgements
We are indebted to the parents of the twins in the Twins Early Development Study (TEDS) for making the study possible. We thank Charles Curtis for constructing the DNA pools used in this study. TEDS is supported by a programme grant from the UK Medical Research Council (Grant no. G0500079); the association scan of reading disability and ability is supported by a grant from the US National Institute of Child Health and Human Development (Grant no. HD49861). The study was approved by the Institute of Psychiatry/South London and Maudsley Research Ethics Committee and appropriate informed consent was obtained from the parents and teachers of the children.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
Supplementary information
Rights and permissions
About this article
Cite this article
Meaburn, E., Harlaar, N., Craig, I. et al. Quantitative trait locus association scan of early reading disability and ability using pooled DNA and 100K SNP microarrays in a sample of 5760 children. Mol Psychiatry 13, 729–740 (2008). https://doi.org/10.1038/sj.mp.4002063
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.mp.4002063
Keywords
This article is cited by
-
Evidence for specificity of polygenic contributions to attainment in English, maths and science during adolescence
Scientific Reports (2021)
-
Genome-wide association scan identifies new variants associated with a cognitive predictor of dyslexia
Translational Psychiatry (2019)
-
Single Nucleotide Polymorphisms Associated with Reading Ability Show Connection to Socio-Economic Outcomes
Behavior Genetics (2017)
-
Decision tree analysis of genetic risk for clinically heterogeneous Alzheimer’s disease
BMC Neurology (2015)
-
The correlation between reading and mathematics ability at age twelve has a substantial genetic component
Nature Communications (2014)