Abstract
Identification of HER2/neu status is important for predicting response to specific chemotherapy in breast carcinoma. Chromogenic in situ hybridization was performed using tissue microarray technology on 188 primary breast carcinomas. To validate the reliability of novel chromogenic in situ hybridization technology, the results of chromogenic in situ hybridization were correlated with the results of two-color fluorescence in situ hybridization done with the same tumors. On tissue microarray panels containing 188 breast carcinoma tissues, fluorescence in situ hybridization and chromogenic in situ hybridization were conducted simultaneously. HER2/neu amplification was detected in 46 tumors (24.5%) by fluorescence in situ hybridization and in 43 tumors (22.9%) by chromogenic in situ hybridization. Results of each method agreed with each other in 177 tumors (concordance: 94.1%). HER2/neu amplification by fluorescence in situ hybridization was associated with nuclear pleomorphism (P = .021), and HER2/neu amplification by chromogenic in situ hybridization was associated with poor nuclear grade (P = .037). High concordance between fluorescence in situ hybridization and chromogenic in situ hybridization indicated that chromogenic in situ hybridization can be a tempting alternative to fluorescence in situ hybridization for the detection of HER2/neu amplification in breast carcinoma because of its accuracy and relative low cost. HER2/neu appeared to have a prognostic implication because its amplification was associated with aggressive biologic features of the breast carcinoma. Integration of tissue microarray technology enabled high-throughput determination of HER2/neu amplification profile with rapidity and accuracy in large cohorts of the breast carcinoma.
Similar content being viewed by others
INTRODUCTION
Among the numerous oncogenes and their products, HER2/neu is the most widely exploited one in clinical oncology. HER2/neu has moved from a laboratory-based prognostic factor to a target for the specific therapy, trastuzumab (Herceptin; Genentech, Inc., South San Francisco, CA), which binds to HER2/neu protein. The HER2/neu is a 185-kDa transmembrane tyrosine kinase, and overexpression of HER2/neu protein arises from HER2/neu gene amplification, resulting in increased gene number (1). HER2/neu overexpression has been shown in 20–40% of human breast carcinomas and is associated with poor clinical outcome, even with systemic chemotherapy (2, 3, 4). Recent studies suggest that HER2/neu is a useful determinant of response to hormonal or cytotoxic chemotherapy. Data from Cancer and Leukemia Group B 8869 and the National Surgical Adjuvant Breast and Bowel Project protocol B-11 suggest that patients whose tumors overexpress HER2/neu may derive a preferential benefit from treatment with doxorubicin (5, 6). With recent introduction of trastuzumab therapy, it is now reasonable to test all new cases of breast carcinomas, both early and late stage, for HER2/neu overexpression. However, there is no consensus about which is the most optimal and accurate testing strategy.
A vast majority of HER2/neu studies has been performed using immunohistochemistry that detects the HER2/neu protein overexpression. Measurement of HER2/neu gene amplification is more accurate because protein overexpression is the result of gene amplification. Introduction of fluorescence in situ hybridization (FISH) allows assessment of the level of gene amplification with information about distribution of gene copies in histologic sections (7). A number of reports have verified its accuracy and apparent superiority over immunohistochemistry in a prediction of response to trastuzumab in metastatic breast carcinoma patients (8, 9). The main difficulty for adopting FISH in a clinical setting is the need of additional equipment for analysis such as fluorescence microscopy and multiband fluorescence filters. Recently, novel technology to detect DNA probe has been developed. Chromogenic in situ hybridization uses a simple immunohistochemistry-like peroxidase reaction (10). Chromogenic in situ hybridization is a tempting technology to overcome the practical limitations of FISH, although its standardization has not been validated.
Completion of the human genome sequence has provided the basic structural information on all human genes. Functional technique such as cDNA microarrays enables analysis of expression levels of thousands of genes and proteins at once (11). Compared with the high-throughput techniques of genomics and proteomics, most tissue-based molecular analyses have been tedious and require extensive manual interaction. Tissue microarray is a novel technology of harvesting small disks of tissue from individual donor paraffin-embedded tissue blocks and placing them in a recipient block with defined array coordinates (12). Tissue microarray technology allows high-throughput molecular profiling of cancer from DNA to protein level by enabling the simultaneous analysis of hundreds of tissue specimens (13). This technology provides maximal use of limited tissue resources and renders the advantage of generating gene expression profiles of cells as they occur in actual neoplastic tissues in vivo. Tissue microarray technology has the potential to significantly accelerate molecular studies and has become one of the most promising tools in cancer research fields.
In this report, we summarize the results of simultaneous analyses for HER2/neu amplification in 188 human breast carcinomas using tissue microarray technology. Chromogenic in situ hybridization appeared as a reasonable alternative to FISH in the current study, and genetic analyses on the archival cancer tissues were successful with novel technologies.
MATERIALS AND METHODS
Materials
One hundred eighty-eight primary breast carcinomas were collected at Inje University Sanggye Paik Hospital, Seoul, Korea. Histopathologic classification and determination of tumor collecting regions were done on hematoxylin and eosin–stained slides. The invasive ductal carcinoma was graded I, II, or III with the Nottingham histologic grading system (14) in ascending degree of malignancy and was graded I, II, or III with the Black's nuclear grading system (15) in descending degree of malignancy.
Tissue Microarray Block
Recipient blocks were made with purified agar in 3.8 × 2.2–cm frames. Holes with 2 mm in each size were made on the recipient blocks by core needle, and agar core was discarded. Donor blocks were prepared after through evaluation of hematoxylin and eosin slides. Representative cancer portions caught from matching donor blocks were transplanted to the recipient blocks using a 2-mm core needle. Recipient blocks were framed in the mold that is used to frame conventional paraffin block, and then paraffin was added to the frame. Consecutive sections in 3.5-μm thickness were cut from the recipient blocks using an adhesive-coated slide system (Instrumedics Inc., New Jersey) supporting the cohesion of the 2-mm array elements on the glass (Fig. 1).
Fluorescence In Situ Hybridization
Two-color FISH was done on a 3.5-μm-thick consecutive microarray sections from the same paraffin blocks with chromogenic in situ hybridization. Before hybridization, the sections were deparaffinized, air dried, and dehydrated in 100% ethanol after incubation at 56° C for 24 hours. Microarray slides were treated in wash buffer (Vysis Inc., Downers Grove, IL) for 3 minutes after treatment with 0.2 n HCl for 20 minutes. Pretreatment solution (Vysis) at 80° C was applied for 30 minutes, and the slides were washed with purified water. Slides were treated with wash buffer twice for 5 minutes serially. Immersed slides in protease solution (Vysis) at 37° C was applied for 10 minutes, and the slides were washed with wash buffer at 45–50° C and air dried. Slides were fixed in 10% buffered formalin for 10 minutes and were washed with wash buffer at 45–50° C. For denaturation, slides were immersed in denaturation solution (Vysis) for 5 minutes at 72° C, followed by dehydration with 70%, 85%, 100% ethanol serially at 45–50° C. For hybridization, 20 μL of LSI HER-2/CEP17 probe (PathVysionTM, Vysis) was applied, and a coverslip was applied over the probe. After overnight hybridization at 37° C in a humidified chamber, the slides were washed with 72° C posthybridization wash buffer (Vysis) for 2 minutes. Nuclei were counterstained with 20 μL 4,6-diamino-2-phenylindole (DAPI) (Vysis). The centromere 17 (CEP) and HER2/neu copy numbers were estimated for the predominant tumor cell population.
Hybridization signals were enumerated by the ratio of orange signals for HER2/neu to green signals for CEP in morphologically intact and nonoverlapping nuclei. At least two times more HER2/neu signals than CEP17 signals in the tumor cells was considered as the criterion for HER2/neu amplification.
Chromogenic In Situ Hybridization
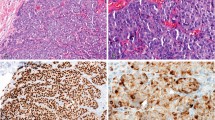
Chromogenic in situ hybridization was done on 3.5-μm-thick tissue microarray sections. Tissue microarray slides were deparaffinized and were incubated in a SPOT-Light Heat Pretreatment buffer (Zymed Inc., South San Francisco, CA) at 92–100° C for 15 minutes. After washing with phosphate buffer saline, 100 μL of SPOT-Light Tissue Pretreatment Enzyme (Zymed) was applied at 37° C for 5 minutes. Microarray slides were washed with phosphate buffered saline and were dehydrated with graded ethanols. A coverslip was applied on microarray slide after application of 15 μL of digoxigenin-labeled HER2/neu probe. The slides were treated on a 95° C hot plate for 5–10 minutes and were incubated at 37° C for 16–24 hours. After incubation, microarray slides were treated in 0.5× sodium chloride citrate for 5 minutes and were washed with phosphate-buffered saline/Tween solution. The slides were treated with 3% hydrogen peroxide for 10 minutes. One hundred μL of FITC-sheep anti-digoxigenin (Zymed) was applied for 30–60 minutes after application of 100 μL of CAS-Block (Zymed) application for 10 minutes. After washing with phosphate buffered saline, 100 μL of HRP-goat anti-FITC (Zymed) was applied for 30–60 minutes. After washing with phosphate buffered saline, 150 μL of 3,3-diaminobenzidine tetrahydrochloride was applied for 20–30 minutes. Microarray slides were counterstained with hematoxylin and eosin after washing with purified water and dehydration with ethanol and xylene.
Amplification of HER2/neu was defined when gene copy number was more than four or when a large gene copy cluster was seen in >50% of cancer cell nuclei.
Data Analysis
Results from chromogenic in situ hybridization and FISH were merged and analyzed. The χ2 test was used for data analysis, and correlation between the results was estimated by Spearman's correlation coefficient (κ). A κ value of 1 denotes complete agreement, values of 0.75 denote excellent agreement, values between 0.4 and 0.75 denote fairly good agreement, and values of <0.4 denote poor agreement.
RESULTS
We performed FISH and chromogenic in situ hybridization simultaneously for the validation of feasibility of chromogenic in situ hybridization in HER2/neu amplification assay. HER2/neu amplification was detected in 46 cases (24.5%) by FISH and in 43 (22.9%) of 188 by chromogenic in situ hybridization (Fig. 2 and Fig. 3). Results of each method agreed with each other in 177 cases (94.1%), whereas results of 11 cases were different from each other. HER2/neu amplification was detected by chromogenic in situ hybridization in 4 cases, although the amplification signals were not observed by FISH in these cases. Results of another 7 cases were vice versa (Table 1). Between the two methods, κ value was 0.838. The results of the study indicated that efficiency of the two methods was equivalent to each other.
Among the 178 invasive ductal carcinoma, 22 (12.4%) were histologic Grade I, and 83 (46.6%) and 73 (41.0%) were Grades II and III, respectively. Seventy-seven (43.3%) were nuclear Grade I, and 84 (47.2%) and 17 (9.6%) were Grades II and III, respectively. HER2/neu amplification by FISH and chromogenic in situ hybridization was associated with poor nuclear grade (P = .043 and P = .037). However, it was not associated with histologic grade. One of the histologic characteristics, nuclear pleomorphism, was associated with HER2/neu amplification by FISH (P = .021). However, it was not associated with HER2/neu amplification by chromogenic in situ hybridization (P = .064). No significant association was found between tubule formation or mitotic counts and HER2/neu amplification (Table 2 and 3).
DISCUSSION
This is the first study that investigated the correlation between chromogenic in situ hybridization and FISH using tissue microarray technology. A study that was conducted using conventional histologic blocks demonstrated that the results of chromogenic in situ hybridization correlated well with those of FISH in archival breast carcinoma samples (10). A significant correlation between chromogenic in situ hybridization and FISH was affirmed in the current study with high concordance rate. High concordance between chromogenic in situ hybridization and FISH for the detection of HER2/neu status in the current study seems to be partly influenced by the advantage of tissue microarray technology. Problems of tissue heterogeneity could be minimized because three assay methods were performed on the consecutive sections of microarray blocks that contain almost the same cancer cells in their biologic states.
The current study indicated that tissue microarray technology was feasible for assaying gene amplification with a limited tissue volume. We used a 2-mm-sized needle for collecting microarray panels. A tissue microarray panel originally was developed that is as small as 0.6 mm in diameter (12). Heterogeneity of the breast carcinoma sometimes makes it difficult to accurately analyze the biologic properties of individual cancers, especially in antigens with heterogeneous staining patterns. Some investigators recommend that punching multiple small cores from different regions captures the heterogeneity of the tumors more effectively (13). We applied a large needle of 2-mm size in collecting the microarray panels to minimize the inadvertent variation in results from tumor heterogeneity. By the time of this writing, three studies have compared biomarker expression using tissue microarrays and conventional histologic sections of the same breast carcinomas (16, 17, 18). Three studies uniformly reported high concordance (90–95%) for expression of biomarkers such as estrogen receptor, progesterone receptor, and HER2/neu. Although the tissue microarray analysis did not represent specific tumor biology completely, tissue microarray analysis had a merit: it could be performed in consecutive sections that had the same cancer tissues in the same coordinate positions as the others.
A main obstacle to the popularization of FISH analysis has been the need to use special fluorescence microscopy with multi-bandpass fluorescence filters that makes it difficult for most institutes to integrate FISH in routine clinical diagnostics. The results of the current study indicated the practical superiority of chromogenic in situ hybridization over FISH in the assessment of gene amplification. Chromogenic in situ hybridization does not require equipment that does not already exist in routine pathologic laboratories. Moreover, most pathologists are familiar with peroxidase-based immunostaining. Another advantage of chromogenic in situ hybridization over FISH in routine practice is that simultaneous verification of histology can be done with chromogenic in situ hybridization. In FISH, sufficient histopathologic evaluation of the individual cells is impossible because of nuclear DAPI staining. The current study indicated that chromogenic in situ hybridization can be used instead of FISH in the screening of HER2/neu amplification in the primary breast carcinomas with feasibility and relative cost-effectiveness.
A commonly expressed concern is whether analysis of molecular targets on tissue microarray may result in lower prevalence than findings obtained from conventional histologic sections. In the results of the current study, prevalence of HER2/neu amplification was 22.9% by chromogenic in situ hybridization and 24.5% by FISH, which well coincides with the results of other studies (7, 8, 9, 10, 19). The validity of tissue microarray analysis has been shown by comparisons with whole-section analysis in breast carcinoma (16, 17, 18). These results together with the current study indicate the feasibility of tissue microarray samples.
The main purpose in analyzing HER2/neu status is to provide the most effective therapeutic regimens for breast cancer patients. Most studies have reported a considerable disagreement in HER2/neu status between FISH and immunohistochemistry analysis (9, 20, 21, 22). Recently, an interesting report has been published (9). The investigators performed FISH and RNA-RNA in situ hybridization for HER2/neu on the same cancer tissues. Results of that study indicated that mRNA expression was highly concordant with FISH and that most cases of immunohistochemistry positive without gene amplification in FISH were devoid of mRNA expression. Hence, those investigators suggested that such cases were most likely false positive and nonspecific. Results from most studies comparing FISH and immunohistochemistry have indicated that FISH was superior to all other methodologies in assessing formalin-fixed, paraffin-embedded material for HER2/neu amplification (10, 11, 12, 13, 14, 15, 16, 17, 18, 19).
Data from the current study indicate that tissue microarray analysis is a feasible and reliable method for assessing HER2/neu amplification with rapidity in a large number of tissues. High concordance of chromogenic in situ hybridization data with those of FISH indicates that chromogenic in situ hybridization can be a tempting alternative to FISH because of its accuracy and relative low cost. Chromogenic in situ hybridization with tissue microarray technology enables high-throughput determination of HER2/ne expression profile and its abnormalities in large cohorts of breast carcinoma. Integration of two novel technologies can provide a rapid validation of identified predictive markers in other cancer research fields.
References
Stern DF, Heffernan PA, Weinberg RA . p185, a product of the neu proto-oncogene, is a receptor-like protein associated with tyrosine kinase activity. Mol Cell Biol 1986; 6: 1729–40.
Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WJ . Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 1987; 235: 177–82.
Paik S, Hazan R, Fisher ER, Sass RE, Fisher B, Redmond C, et al. Pathologic findings from the National Surgical Adjuvant Breast and Bowel Project: prognostic significance of erbB-2 protein overexpression in primary breast cancer. J Clin Oncol 1990; 8: 103–12.
Toikkanen S, Helin H, Isola J, Joensuu H . Prognostic significance of HER-2 oncoprotein expression in breast cancer. A 30-yr follow-up. J Clin Oncol 1992; 10: 1044–8.
Paik S, Bryant J, Park C, Fisher B, Tan-Chiu E, Hyams D, et al. erbB-2 and response to doxorubicin in patients with axillary lymph node-positive, hormone receptor-negative breast cancer. J Natl Cancer Inst 1998; 90: 1361–70.
Muss HB, Thor AD, Berry DA, Kute T, Liu ET, Koerner F, et al. c-erbB-2 expression and response to adjuvant therapy in women with node-positive early breast cancer. N Engl J Med 1994; 330: 1260–6.
Kallioniemi OP, Kallioniemi A, Kurisu W, Thor A, Chen LG, Smith HS, et al. ERBB2 amplification in breast cancer analyzed by fluorescence in situ hybridization. Proc Natl Acad Sci U S A 1992; 89: 5321–5.
Press MF, Bernstein L, Thomas PA, Meisner LF, Zhou JY, Ma Y, et al. HER-2/neu gene amplification characterized by fluorescence in situ hybridization: poor prognosis in node-negative breast carcinomas. J Clin Oncol 1997; 15: 2894–904.
Tubbs RR, Pettay JD, Roche PC, Stoler MH, Jenkins RB, Grogan TM . Discrepancies in clinical laboratory testing of eligibility for trastuzumab therapy: apparent immunohistochemical false-positive do not get the message. J Clin Oncol 2001; 19: 2714–21.
Tanner M, Gancberg D, Di Leo A, Lasimont D, Rouas G, Piccart M, et al. Chromogenic in situ hybridization. A practical alternative for fluorescence in situ hybridization to detect HER-2/neu oncogene amplification in archival breast cancer samples. Am J Pathol 2000; 157: 1467–72.
DeRisi J, Penland L, Brown PO, Bittner ML, Meltzer PS, Ray M, et al. Use of a cDNA microarray to analyze gene expression patterns in human cancer. Nat Genet 1996; 14: 457–60.
Kononen J, Bubendorf L, Kallioniemi A, Barlund M, Schraml P, Leighton S, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med 1998; 4: 844–7.
Kallioniemi OP, Wagner U, Kononen J, Sauter G . Tissue microarray technology for high-throughput molecular profiling of cancer. Hum Mol Genet 2001; 10: 657–62.
Elston CW, Ellis IO . Pathological prognostic factors in breast cancer: I. The value of histologic grade in breast cancer—experience from a large study with long-term follow-up. Histopathology 1991; 19: 403–10.
Black MM, Speeder FD . Nuclear structure in cancer tissue. Surg Gynecol 1957; 105: 97–102.
Bucher C, Torhorst J, Bubendorf L, Schraml P, Kononen J, Moch H, et al. Tissue microarrays (“tissue chips”) for high-throughput cancer genetics: Linking molecular changes to clinical endpoints. Am J Hum Genet 1999; 65 (Suppl): 43.
Gillett CE, Springall RJ, Barnes DM, Hanby AM . Multiple tissue core arrays in histopathology research: a validation study. J Pathol 2000; 192: 549–53.
Camp RL, Charette LA, Rimm DL . Validation of tissue microarray technology in breast carcinoma. Lab Invest 2000; 80: 1943–9.
Pauletti G, Godolphin W, Press MF, Slamon DJ . Detection and quantitation of HER-2/neu gene amplification in human breast cancer archival material using fluorescence in situ hybridization. Oncogene 1996; 13: 63–72.
Press MF, Hung G, Godolphin W, Slamon DJ . Sensitivity of HER-2/neu antibodies in archival tissue samples: potential source of error in immunohistochemical studies of oncogene expression. Cancer Res 1994; 54: 2771–7.
Roche PC, Ingle JN . Increased Her-2 with US Food and Drug Administration approved antibody. J Clin Oncol 1999; 17: 434–5.
Jacobs TW, Gown AM, Yaziji H, Barnes MJ, Schnitt SJ . Specificity of HercepTest in determining Her-2/neu status of breast cancer using the United States Food and Drug Administration-approved scoring system. J Clin Oncol 1999; 17: 1983–7.
Acknowledgements
Supported by Grant R04-2001-00036 from the Korea Science & Engineering Foundation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Park, K., Kim, J., Lim, S. et al. Comparing Fluorescence In Situ Hybridization and Chromogenic In Situ Hybridization Methods to Determine the HER2/neu Status in Primary Breast Carcinoma using Tissue Microarray. Mod Pathol 16, 937–943 (2003). https://doi.org/10.1097/01.MP.0000086487.78558.7D
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1097/01.MP.0000086487.78558.7D
Keywords
This article is cited by
-
Decreased expression of C-erbB-2 and CXCR4 in breast cancer after primary chemotherapy
Journal of Translational Medicine (2012)
-
Chromogenic in-situ hybridization: a viable alternative to fluorescence in-situ hybridization in the HER2 testing algorithm
Modern Pathology (2006)
-
Topoisomerase II-alpha gene deletion is not frequent as its amplification in breast cancer
Breast Cancer Research and Treatment (2006)
-
Reliability of chromogenic in situ hybridization for detecting HER-2 gene status in breast cancer: comparison with fluorescence in situ hybridization and assessment of interobserver reproducibility
Modern Pathology (2005)
-
Chromogenic in situ hybridization (CISH): a novel alternative in screening archival breast cancer tissue samples for HER-2/neu status
Breast Cancer Research (2004)