Abstract
Purpose: To compare the efficacy of chromogenic in situ hybridization (CISH™) with fluorescence in situ (FISH) hybridization and immunohistochemistry (IHC) in determination of the HER2 status in human breast cancer. Materials and Methods: HER2 gene amplification was determined on formalin-fixed paraffin-embedded (FFPE) sections of 62 invasive breast cancers by FISH and followed by CISH using a digoxigenin (DIG)-labeled HER2 DNA probe generated by Subtraction Probe Technology (SPT™), and a biotin-labeled chromosome 17 centromeric (chr.17cen) probe. The sections were heat treated and enzyme digested. After in situ hybridization, the HER2 probe was detected with fluorescein (FITC)-anti-DIG for FISH, followed by peroxidase-anti-FITC and diaminobenzidine (DAB) for CISH. The chr.17cen probe was detected with peroxidase–streptavidin and DAB. For CISH application, HER2 gene copies or chromosome 17 centromeres and morphology of cells were easily visualized simultaneously with a 40 × objective under bright-field microscope in hematoxylin-counterstained sections. IHC study of HER2 overexpression was performed on adjacent sections using a panel of three HER2 antibodies (TAB 250, CB11, A0485), and staining was scored according to the criteria specified in the HercepTest. Results: HER2 gene amplification detected by CISH was visualized typically as large DAB-stained clusters or by many dots in the nucleus. FISH and CISH identified HER2 gene amplification in 19% of the tumors. Chromosome 17 polysomy was detected in 31% of the tumors. HER2 overexpression was demonstrated in 19% (TAB 250), 23% (CB11), and 36% (A0485) of the tumors. Complete concordance between the results of CISH with FISH, TAB 250, CB11, and A0485 was seen in 100%, 97%, 94%, and 84% of the cases, respectively. Conclusion: By permitting observation of morphology using a bright-field microscope, CISH is an accurate, practical, and economical approach to screen HER2 status in breast cancers. It is a useful methodology for confirming ambiguous IHC results.
Similar content being viewed by others
INTRODUCTION
HER2 gene amplification or HER2 protein overexpression has been identified in 10–34% of invasive breast cancers according to a series of 52 published studies including >16,000 patients and using different methodologies (1). Identification of HER2 status is important for determining the prognosis of patients who have invasive breast cancer, as well as for selecting a subgroup of them with metastasis HER2 overexpression for therapy with trastuzumab (Herceptin; Genentech, Inc., South San Francisco, CA) (2, 3), a monoclonal antibody to the HER2 protein. Trastuzumab has been found to be effective only in patients whose tumors show HER2 gene amplification and/or HER2 protein overexpression. Therefore, accurate, consistent, and straightforward methods for evaluation of HER2 status have become increasingly important.
IHC staining has been the predominant method of determining HER2 status in breast cancer specimens. It is relatively easy to perform and has rapid turnaround time, and relatively low cost (1, 4). However, commercially available antibodies have demonstrated wide variation in sensitivity and specificity for FFPE tissue samples, and the effect of the tissue fixative and pretreatment have a substantial effect on HER2 IHC staining (1, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21), although antigen retrieval and immunodetection system currently used have improved sensitivity of antibodies. In addition, the lack of universal scoring system and interobserver differences in interpretation of HER2 IHC result are also important issues (1, 15, 17, 18, 22, 23).
Overexpression of the HER2 protein generally (>95%) results from HER2 gene amplification (20, 24, 25, 26). Fluorescence in situ hybridization (FISH) is thought to be the most sensitive technique for quantitative evaluation of HER2 gene status in breast cancer cells, and FISH in FFPE tissue sections is a valid alternative to IHC (1, 24, 27). In a recent study, the sensitivity and specificity of FISH were superior to IHC in FFPE tissue sections (10). Patients who were positive by FISH but negative by IHC had a worse survival than did those who had HER2 overexpression but absence of gene amplification (10). Therefore, HER2 amplification might provide more meaningful prognostic information than HER2 overexpression in breast cancer patients. In addition, FISH quantifies the number of gene copies in the cancer cell, which objectively reflects the HER2 gene status of tumors, whereas IHC is a more subjective test. Therefore, FISH can be easier to interpret than IHC. However, FISH methodology also has its disadvantages. Evaluation of FISH requires a modern and expensive fluorescence microscope equipped with high-quality 60 × or 100 × oil immersion objectives and multi–band-pass fluorescence filters, which is not used in most routine diagnostic laboratories; The fluorescence signals can fade within several weeks, and the hybridization results are typically recorded with an expensive CCD camera. Therefore, analysis and recording of FISH data is expensive and time consuming. Most important, tissue section morphology is not optimal in FISH on FFPE, a particular problem for distinguishing invasive breast cancer and breast carcinoma in situ, where HER2 gene amplification or protein overexpression has different clinical significance. All of these limitations make FFPE FISH cumbersome for routine work (6, 28).
To accurately identify HER2 gene status by bright-field microscopy and overcome FISH limitations, we have developed CISH using the HER2 DNA probe generated by Subtracted Probe Technology (SPT). With SPT, repetitive DNA sequences, such as Alu and LINE elements, which may consist of up to 40% of template and cause unspecific hybridization, are quantitatively removed. Therefore, the final probe is very specific, and the need for blocking nonspecific hybridization with Cot-1 DNA in traditional FISH probes is eliminated. After hybridization of the probe to target DNA on the tissue section, the DIG-labeled DNA probe is detected first by FISH using FITC-conjugated anti-DIG antibody and then followed by CISH using HRP conjugated anti FITC antibody and a simple IHC-like peroxidase reaction. Further, a chr.17cen probe was used as a reference probe to assist in distinguishing HER2 amplification from chromosomal polysomy. CISH using the SPT HER2 DNA probe has already been evaluated against FISH (Vysis) and a HER2 antibody CB11 (28). The conclusion of that study was that CISH is a useful alternative for detection of HER2 amplification in FFPE tumor samples, especially for confirming the IHC staining results. To further evaluate and assess the clinical usefulness of CISH using SPT™ HER2 probe, three commonly used and commercially available HER2 antibodies: monoclonal TAB 250 and CB11, which recognize the external and internal domains of HER2 oncoprotein, respectively, and A0485, a rabbit polyclonal antibody which recognizes internal domains, were compared in this study with CISH. As no internationally accepted standard exists, we applied the scoring proposed by the HercepTest (DAKO) for the interpretation of the immunoreactivity of these three antibodies.
MATERIALS AND METHODS
Tumor Specimens
FFPE tissue blocks from 62 patients with invasive breast cancer were studied. Histopathological classification was done on hematoxylin and eosin (H&E)-stained slides according to standard histopathological practice.
FISH and CISH
Probes
HER2 gene status was determined by using DIG labeled SPOT-Light® HER2 DNA probe (Zymed, South San Francisco, CA). Biotin-labeled chr.17cen probe (Zymed) was used for the tumors having more than two HER2 gene copies per nucleus (34 of 62 cases).
FISH and CISH procedure
FISH and CISH was done on 4 μm-thick tissue sections mounted on Superfrost/plus microscope slides (Fisher, Pittsburgh, PA). The slides were baked for 2 to 4 hours at 65° C and then deparaffinized 10 minutes in xylene twice, and 5 minutes in ethanol, thrice. Air-dried tissue sections were placed in a plastic Coplin jar containing the CISH Pretreatment Buffer (SPOT-Light® Tissue Pretreatment Kit, Zymed), and loosely capped. They were heated at 92° C for 15 minutes in the microwave with a temperature probe (GE Profile Sensor convection). The temperature probe was placed in a separate plastic Coplin jar without a cap. The slides were washed immediately with deionized water after heat pretreatment. Enzyme digestion was followed by covering the section with prewarmed 37° C pepsin (SPOT-Light® Tissue Pretreatment Kit, Zymed) and by incubating at 37° C for 3 ± 1 minutes. The slides were then washed with deionized water, dehydrated with graded ethanol, and air-dried. The ready-to-use DIG-labeled HER2 probe or biotin-labeled chr.17cen probe was applied to the center of the coverslip. The coverslip was placed with probe side down on the tissue sample. Fifteen to 20 μL of the probe was used for 22 × 22-mm or 24 × 32-mm coverslips according to the size of the tissue sections to be covered. After sealing the edges of the coverslips with rubber cement, the tissue sections and the probes were denatured at 94° C for 5 minutes by placing the slides in the slide block of the PCR machine (MJ Research, Watertown, MA). Hybridization was done in the same slide block at 37° C overnight. The stringent wash was done with 0.5 × standard saline citrate at 75 to 80° C for 5 minutes. The following steps with CISH Detection Kit (Zymed) are similar to the case of IHC. The endogenous peroxidase activities were blocked in 3% H2O2 diluted with methanol for 10 minutes. The unspecific staining was blocked by applying the Cas-Block on the section and by incubating for 10 minutes. Followed an incubation with FITC conjugated mouse anti-DIG antibody for 45 minutes at room temperature the HER-2 probe was detected by FISH. After FISH evaluation, the mounting medium with DAPI (VECTASHIELD, Vector Laboratories, Inc., Burlingame, CA) was washed away with PBS and Tween20. CISH procedure was continued by incubation with HRP conjugated sheep anti FITC antibody for 45 minutes at room temperature, followed by DAB development for 30 minutes. The biotin-labeled chr.17cen probe was detected with sequential incubation with HRP-conjugated streptavidin for 45 minutes at room temperature and DAB development (CISH Centomere Detection Kit, Zymed) for 30 minutes. Tissue sections were counterstained with hematoxylin, dehydrated, and coverslipped. Positive controls were included in each staining run.
Evaluation of FISH and CISH Results
The FISH results were evaluated using a microscope (Leica DMLB) equipped with 10 ×, 20 ×, and 40 × dry objectives and 100 × oil immersion objective; filter sets of DAPI/FITC/TRITC (Triple band), TRITC, and FITC (Chroma Technology Corp, Brattleboro, VT); and SPOT CCD camera (Diagnostic Instruments, Inc., Sterling Heights, MI). CISH results were evaluated using a microscope (Nikon, E400) equipped with 10 ×, 20 ×, and 40 × dry objectives, with 10 × oculars. Revised standard, as described in Tanner's paper (28), was used to interpret the HER2 FISH and CISH results (Table 1).
IHC
HER2 antibodies
Two monoclonal antibodies (TAB 250, Zymed; CB11, Novocastra, Newcastle, United Kingdom) and one polyclonal antibody (A0485, DAKO, Carpinteria, CA, same antibody used in the HercepTest) were tested for HER2 immunostaining on a serial of 4-μm sections for the 62 tumors. Each of these antibodies has been reported to specifically recognize the HER2 receptor protein either in published literature or in commercial information.
IHC procedure
Tissue sections were deparaffinized followed by antigen retrieval in citrate buffer (pH 6.0, Zymed) for CB11 and A0485, or by Ficin (Digest All I, Zymed) pretreatment for 10 minutes at 37° C for TAB 250. After blocking for non-specific antibody binding (NBA kit, Zymed), primary antibody was added on the section and incubated for 1 hour at room temperature. The NBA Kit (Non-biotin amplification system, Zymed) was used for visualization with DAB as the chromogen. The sections were counterstained with hematoxylin.
Evaluation of IHC results
For determination of HER2 protein overexpression, only the membrane-staining pattern and staining intensity of invasive tumor cells were scored. Interpretation was independently performed by two of the authors (JZ and RW), each blinded against the other and to the results of the other assays. For the three antibodies used in this study, IHC staining was scored according to the criteria specified by DAKO for the interpretation of the HercepTest. Immunoreaction was determined to be strongly positive (3+) if a strong complete membrane staining was observed in >10% of the tumor cells or to be weakly positive (2+) if >10% of the tumor cells showed weak to moderate complete membrane staining. All other staining patterns were interpreted as negative (0/+1). The investigators generally came to similar estimates of the stained cells on the slides and to identical scoring results. If there was any disagreement, a third person's (Z.S.) interpretation was included.
RESULTS
FISH and CISH
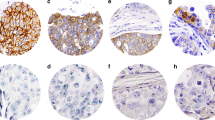
Criteria for successful FISH or CISH analysis included identification of at least one copy of the HER2 gene per nucleus in most cancer cells and appropriate pepsin digestion as evidenced by well-preserved cell morphology. FISH and CISH were successful in 85% of cases in the first run. In the end, FISH and CISH for HER2 was successful in all 62 cases and for chr.17cen was successful for all cases attempted (total 34 cases). Pepsin digestion was adjusted to 10 sec for one case and to 3 minutes with 10 × pepsin for another case. The FISH signals were seen by 40 × objective and were easily identified by 100 × objective. The CISH staining results were clearly seen using a 40 × objective in tissue sections that were counterstained with hematoxylin. Tumors with HER2 gene amplification appeared typically as large peroxidase-positive intranuclear gene copy clusters (Fig. 1A), as numerous individual peroxidase-positive small signals (Fig. 1B), or as a mixture of clusters and individual gene copies (Fig. 2A). Tumors with normal HER2 gene showed typically one to two dots per nucleus (Fig. 2C). Tumors with HER2 low amplification showed typically 6 to 10 gene copies per nucleus (Fig. 3A), which was confirmed by CISH using chr.17cen probe on an adjacent section (Fig. 3B). Tumors with polysomy showed typically three to five HER2 gene copies per nucleus (Fig. 3D), which was also confirmed by CISH using chr.17cen probe on an adjacent section (Fig. 3E).
HER2 CISH and IHC staining in breast carcinomas. CISH of HER2 gene amplification appears as a mixture of clusters and individual gene copies (A), and IHC of TAB250 3+ staining (B); CISH of normal HER2 gene appears as one or two gene copies (C), and IHC of TAB250 0 staining (D). Original magnification, 400 ×. Counterstained with hematoxylin.
HER2 and Chr.17cen CISH, and HER2 IHC staining in breast carcinomas. CISH of HER2 gene low amplification appears as 6 to 10 gene copies per nucleus (A), CISH using Chr.17cen probe showing diploid of chromosome 17 as two dots per nucleus (B), and IHC of TAB250 2+ staining (C); CISH of HER2 appears as three to five copies per nucleus (D), CISH using Chr.17cen probe showing polysomy of chromosome 17 as three to five dots per nucleus (E), and IHC of TAB250 0 staining (F). Original magnification, 400 ×. Counterstained with hematoxylin.
In a series of 62 breast cancers, the prevalence of HER2 amplification was 19% (12 of 62) by FISH and CISH (Table 2). Among the 12 HER2 amplification cases, 1 was HER2 low amplification (6–10 gene copies per nucleus for >50% of tumor cells).
Of 62 tumors tested by HER2 CISH, 28 tumors had one to two HER2 gene copies/nucleus in tumor cells. Because there have been no previous reports of HER2 gene deletion, we consider these 28 tumors not to have polysomy of chromosome 17. Chr.17cen CISH was tested in the remaining 34 tumors, and chromosome 17 polysomy was found in 19 tumors. Therefore, polysomy of chromosome 17 in current study is 31% (19/62). Eight of 12 tumors (67%) with HER2 amplification were chromosome 17 polysomy. Most of the tumors having chromosome 17 polysomy showed three to five dots per nucleus. However, one case (Case 59, Table 3) showed four to seven dots per nucleus. Perhaps it was due to the presence of numerous cycling cells at S and G2 phases of the cell cycle. Another case (Case 53, Table 3) had three to five HER2 gene copies per nucleus but had one to two dots per nucleus for chr.17cen probe. We think this corresponded to has sometimes been seen by CGH of duplication of chromosome arm 17q (29 and personal communication with Dr. Jorma Isola). Therefore, Case 53 was not classified as HER2 amplification.
The concordance between FISH and CISH was 100%.
IHC
HER2 protein high overexpression (3+) was clearly seen using 10 × or 20 × objectives in tissue sections that were counterstained with hematoxylin (Fig. 2B). HER2 protein weak overexpression (2+) was clearly distinguishable using 20 × and 40 × objectives (Fig. 3C). HER2 protein no overexpression (0, 1+) was distinguished using 20 × and 40 × objectives (Fig. 2D, 3F).
In a series of 62 breast cancers, the prevalence of HER2 protein overexpression was 19% by TAB 250, 23% by CB11, and 36% by A0485 (Table 2). There was a 97% concordance between the results obtained with the TAB 250 and CB11 antibodies. The concordance between the results obtained with TAB 250 and A0485 or CB11 and A0485 is both 86%.
Correlation of CISH with IHC
The relationship between gene amplification and protein overexpression of HER2 is illustrated in Table 2.
The results of TAB 250 and CISH were discordant in two cases (Cases 10 and 24, Table 3): Case 10 had HER2 gene amplification but was negative by TAB 250; Case 24 did not have HER2 gene amplification but was 2+ by TAB 250. The results of CB11 and CISH were discordant in four cases: Case 10 had HER2 gene amplification but was negative for CB11; the other three cases (Case 24 and two other cases which were not listed in Table 3) did not have HER2 gene amplification but were 2+ by CB11. Two of these 3 cases had chromosome 17 polysomy. The results of A0485 and CISH were discordant in ten cases: all the cases had overexpression of the protein but were absent of gene amplification (Case 24, 53,59 and other seven cases were not listed in Table 3). Eight of these 10 cases had chromosome 17 polysomy.
With one exception (Case 10 in Table 3), 11 of 12 cases with HER-2 gene amplification detected by CISH definitely showed positive staining by all three antibodies. With two exceptions (Cases 10 and 24), all the cases with 3+ positive for A0485 had HER2 amplification. Six of eight (75%) cases with 2+ positive by A0485 were negative by TAB 250 as well as by CB11, and were chromosome 17 polysomy lacked HER2 amplification. The concordance between the results of TAB 250 and CISH, CB11 and CISH, and A0485 and CISH were 97%, 94%, and 84%, respectively.
DISCUSSION
For routinely used HER2 testing, accuracy and ease are essential for starting trastuzumab (Herceptin) therapy for metastasis breast cancer. IHC and FISH on FFPE tissue are the two methodologies currently FDA approved for use in HER2 testing. In our 62 cases, results of FISH and CISH have 100% agreement. The current version of HER2 FISH is based on single-color detection, which is similar as INFORM FISH test (Ventana, Tucson, AZ). The HER2 CISH is a continuation of FISH with IHC like reaction. The complete agreement of FISH and CISH results demonstrated that HER2 CISH is as sensitive as HER2 FISH. The present study, together with those of a previous article (28), demonstrated the utility of CISH, a novel methodology, in the determination of HER2 amplification in FFPE tumor samples. Interpretation of CISH is performed using a standard light microscope and permits simultaneous evaluation of gene copies, tumor cell and surrounding tissue morphology on the same slide. By contrast, tissue morphology is less optimal with FISH, and it may be necessary to review H&E-stained sections to definitely identify areas of invasive carcinoma from carcinoma in situ. This is a disadvantage of FISH methodology in routine pathology. In addition, FISH takes much longer time to analyze than CISH, and FISH slides must be stored at 4° C or lower and are subject to quenching of the fluorescent signal, whereas CISH-stained slides can be stored in standard slide files and the reaction product is permanent.
The current HER2 CISH is based on single-color detection. To avoid misinterpretation of gene amplification with polysomy, we used the chr.17cen probe on adjacent sections of the tumors which have more than two HER2 gene copies per nucleus. The increased HER2 copy number was evaluated both as absolute numbers of HER2 or as clusters of dots per nucleus, and as HER2 copy numbers relative to chr.17cen copy numbers. By using the HER2 probe and chr.17cen probe on serial sections, simultaneous detection of oncogene and chromosome copy numbers was ascertained. To evaluate HER2 CISH, we defined amplification as clusters of dots or as more than 6 gene copies per nucleus in >50% of invasive tumor cells; polysomy as three to five gene copies per nucleus in >50% of invasive tumor cells; and normal as HER2 CISH showing 1–2 gene copies per nucleus in >50% of invasive tumor cells. The chr.17cen probe was applied to the tumors that had more than two gene copies per nucleus. It was only informative for Case 59 (Table 3), which had four to seven HER2 gene copies per nucleus, because the use of the centromeric probe confirmed that it was due to chromosome 17 polysomy. Although chr.17cen probe confirmed that the tumor with 6–10 HER2 gene copies per nucleus was due to HER2 gene low amplification and the tumors (total 12) with 3–5 HER2 gene copies per nucleus were due to chromosome 17 polysomy, chr.17cen probe did not provide more information for these tumors. As it has been suggested by Tubbs (30), we suggest that the chr.17cen probe is only necessary when there are 6–10 HER2 gene copies per nucleus to confirm HER2 gene amplification, which is relatively rare (one out of 62 cases, 1.6%). We do not think that 2–4 HER2 gene copies per nucleus can even be due to gene amplification. In the invasive breast cancers we studied, chromosome 17 polysomy was frequent (31%), which is similar to the results reported by others (13, 24, 31) and even more frequent (67%) in the invasive breast cancers with HER2 amplification.
In term of successful rate of CISH, we found that the heat pretreatment and pepsin digestion are the most critical procedures for successful and optimized CISH performance. We have also observed from recent experiment that successful rate of CISH was low when over 20-year tissue blocks were used. In a recent study (42) the authors have employed CISH for HER2 assessment, in which 2 of 20 cases examined could not yield interpretable CISH. We suspect that pepsin digestion was not optimized for these two cases since pepsin procedure was not described in the paper.
In the current study, correlations between HER2 CISH with 2+ and 3+ cases of TAB 250, CB11, and 3+ cases of A0485 on invasive breast cancers using FFPE tissue were generally good, and are similar to that seen in prior studies comparing assays of HER2 gene amplification with those of HER2 protein expression (6, 20, 24, 25, 26, 32, 33). Our study showed that TAB 250 had the lowest misclassification rate compared with CISH result. All tumors except one (Case 10) with HER2 gene amplification demonstrated overexpression of HER2 protein with the three antibodies used in this study. This outlying case may present a small-undetermined percentage that amplified HER2 without overexpression (6, 16, 20, 26). The high rate of concordant results obtained by the two monoclonal antibodies was shown in this study. The monoclonal antibodies detected HER2 overexpression in the absence of gene amplification in 2% (1 of 50, TAB 250) to 6% (3 of 50, CB11) of the cases. These results are in agreement with a published range of other studies and could represent single-copy overexpression at the transcriptional level and/or beyond (6, 10, 20, 24, 25, 26, 33). Alternatively, it may be due to gene amplification that is below the detection level of FISH and CISH. Compared with the monoclonal antibodies, the high level of overexpression of 36% (22 of 62) was detected by the polyclonal antibody A0485, which is in higher range of 10 to 34% HER2 overexpression reported in the literature (1). Although the HercepTest kit was not used in the current study, the same polyclonal DAKO antibody provided in the kit with standard heat induced epitope retrieval was applied. Among the tumors with HER2 overexpression detected by A0485, 36% (8 of 22) did not have HER2 gene amplification and were negative by TAB 250 and CB11. These discrepancies were mainly represented by tumors detected as 2+ by A0485. This observation is in agreement with the recent findings of Ridolfi (11) and Lebeau (16). When closely investigating the tumors stained 2+ by A0485, 6 of 8 (75%) cases with chromosome 17 polysomy lacked HER2 gene amplification. The present study demonstrated again that A0485 has higher sensitivity than other commercially available antibodies, and it might detect lower levels of protein expression as suggested by others (5, 16, 34, 35). It has also been reported by several investigators in independent laboratory facilities that the HercepTest or A0485 results in significant false-positive cases (5, 9, 10, 35, 36, 37). As discussed by Pauletti (10), in a subgroup analysis of the phase III clinical trials that led to approval of trastuzumab (34), patients with 2+ IHC score did not seem to benefit significantly from trastuzumab therapy (38, 39, 40), and the beneficial treatment effects were largely limited to patients with the highest levels of HER2 protein overexpression (3+). In addition, the study has shown that patients positive by HercepTest but negative by TAB 250 had the same response rate as patients who were TAB 250 negative overall (41). Some investigators suggested a revised scoring system by subtracting the level of staining of nonneoplastic epithelium to improve HercepTest specificity (5). However, many tumor specimens in our series did not contain nonneoplastic epithelium, therefore we could not apply this scheme in current study. A major advantage of CISH and IHC as an in situ-based technology is their ability to combine molecular diagnosis with histological examination of the tissue. A combinatorial strategy using IHC and CISH should provide comprehensive and valuable information on both HER2 protein concentrations and gene amplification to help clinicians make crucial management decisions.
In summary, we have documented a complete agreement between CISH and FISH, and a high level of concordance between CISH and IHC, especially between CISH and monoclonal antibody TAB 250, in the evaluation of HER2 status on invasive breast carcinoma. Sixty-seven percent of the tumors that were scored as 2+ by polyclonal antibody A0485 were negative by TAB 250 and CB11, and did not show HER2 gene amplification but showed chromosome 17 polysomy. In agreement with the study by Tanner (28), the present study confirmed the validity of CISH methodology. If such consistency can be reproduced in other laboratories, CISH could prove to be truly valuable in clinical practice.
References
Ross JS, Fletcher JA . HER-2/neu (c-erb-B2) gene and protein in breast cancer. Am J Clin Pathol 1999; 112: S53–S67.
Shak S . Overview of the trastuzumab (Herceptin) anti-HER2 monoclonal antibody clinical program in HER2-overexpressing metastatic breast cancer. Herceptin multinational investigator study group. Cancer Res 1999; 6: 71–77.
Cobleigh MA, Vogel CL, Tripathy D, Robert NJ, Scholl S, Fehrenbacher L, et al. Multinational study of the efficacy and safety of humanized anti-HER-2 overexpression metastatic breast cancer that has progressed after chemotherapy for metastatic disease. J Clin Oncol 1999; 17: 2639–2648.
Hanna W, Kahn HJ, Trudeau M . Evaluation of HER-2/neu (erbB-2) status in breast cancer: from bench to bedside. Mod Pathol 1999; 12: 827–834.
Jacobs TW, Gown AM, Yaziji H, Barnes MJ, Schnitt SJ . Specificity of Hercep Test in determining HER-2/neu status of breast cancers using the United States Food and Drug Administration-approved scoring system. J Clin Oncol 1999; 17: 1983–1987.
Jacobs TW, Gown AM, Yaziji H, Barnes MJ, Schnitt SJ . Comparison of fluorescence in situ hybridization and immunohistochemistry for the evaluation of HER-2/neu in breast cancer. J Clin Oncol 1999; 17: 1974–1982.
Mitchell MS, Press MF . The role of immunohistochemistry and fluorescence in situ hybridization for HER-2/neu in assessing the prognosis of breast cancer. Semin Oncol 1999; 26: 108–116.
Espinoza F, Anguiano A, Roche PC, Ingle JN . The HercepTest assay: another perspective. J Clin Oncol 1999; 17: 2293B.
Jimenez RE, Wallis T, Tabasczka P, Visscher DW . Determination of Her-2/neu status in breast carcinoma: comparative analysis of immunohistochemistry and fluorescent in situ hybridization. Mod Pathol 2000; 13: 37–45.
Pauletti G, Dandekar S, Rong HM, Ramos L, Peng HJ, Seshadri R, et al. Assessment of methods for tissue-based detection of the HER-2/neu alteration in human breast cancer: a direct comparison of fluorescence in situ hybridization and immunohistochemistry. J Clin Oncol 2000; 18: 3651–3664.
Ridolfi RL, Jamehdor MR, Arber JM . HER-2/neu testing in breast carcinoma: a combined immunohistochemical and fluorescence in situ hybridization approach. Mod Pathol 2000; 13: 866–873.
Kakar S, Puangsuvan N, Stevens JM, Serenas R, Mangan G, Sahai S, et al. HER-2/neu assessment in breast cancer by immunohistochemistry and fluorescence in situ hybridization: comparison of results and correlation with survival. Mol Diagn 2000; 5: 199–207.
Gancberg D, Lespagnard L, Rouas G, Paesmans M, Piccart M, Di Leo A, et al. Sensitivity of HER-2/neu antibodies in archival tissue samples of invasive breast carcinomas. Am J Clin Pathol 2000; 113: 675–682.
Vang R, Cooley LD, Harrison WR, Tommy R, Abrams J . Immunohistochemical determination of HER-2/neu expression in invasive breast carcinoma. Am J Clin Pathol 2000; 113: 669–674.
Allred DC, Swanson PE . Testing for erbB-2 by immunohistochemistry in breast cancer. Am J Clin Pathol 2000; 113: 171–175.
Lebeau A, Deimling D, Kalz C, Sendelhofert A, Iff A, Luthardt B, et al. HER-2/neu analysis in archival tissue samples of human breast cancer: comparison of immunohistochemistry and fluorescence in situ hybridization. J Clin Oncol 2001; 19: 354–363.
Press MF, Hung G, Godolphin W, Slamon DJ . Sensitivity of HER-2/neu antibodies in archival tissue samples: potential source of error in immunohistochemical studies of oncogene expression. Cancer Res 1994; 4: 2771–2777.
Busmanis I, Feleppa F, Jones A, McGrath KM, Reed R, Collins J, et al. Analysis of cerbB2 expression using a panel of 6 commercially available antibodies. Pathology 1994; 26: 261–267.
Slamon DJ . Studies of the HER-2/neu proto-oncogene in human breast cancer. Cancer Invest 1990; 8: 253.
Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science 1989; 244: 707–712.
Penault-Llorca F, Adelaide J, Houvenaeghel G, et al. Optimization of immunohistochemical detection of erbB2 in human breast cancer: impact of fixation. J Pathol 1994; 173: 65–75.
Kay EW, Walsh CJ, Cassidy M, Hassoun J, Birnbaum D, Jacquemier J . C-erbB-2 immunostaining: problems with interpretation. J Clin Pathol 1994; 47: 816–822.
Ross JS, Fletcher JA . The HER-2/neu oncoprotein in breast cancer: prognostic factor, predictive factor, and target for therapy. Oncology 1998; 3: 237–252.
Pauletti G, Godolphin W, Press MF, Slamon DJ . Detection and quantitation of HER-2/neu gene amplification in human breast cancer archival material using fluorescence in situ hybridization. Oncogene 1996; 13: 63–72.
Kallioniemi O-P, Kallioniemi A, Kurisu W, Thor A, Chen LC, Smith HS, et al. ERBB2 amplification in breast cancer analyzed by fluorescence in situ hybridization. Proc Natl Acad Sci U S A 1992; 89: 5321–5325.
Ciocca DR, Fujimura FK, Tandon AK, Clark GM, Mark C, Lee-Chen GJ, et al. Correlation of HER-2/neu amplification with expression and with other prognostic factors in 1103 breast cancers. J Natl Cancer Inst 1992; 84: 1279–1282.
Press MF, Bernstein L, Thomas PA, Meisner LF, Zhou JY, Ma Y, et al. HER-2/neu gene amplification characterized by fluorescent in situ hybridization: poor prognosis in node-negative breast carcinoma. J Clin Oncol 1997; 15: 2894–2904.
Tanner M, Gancberg D, Di Leo A, Larsimont D, Rouas G, Piccart MJ, et al. Chromogenic in situ hybridization: a practical alternative for fluorescence in situ hybridization to detect HER-2/neu oncogene amplification in archival breast cancer samples. Am J Pathol 2000; 157: 1467–1472.
Isola J, Chu L, DeVries S, Matsumura K, Chew K, Ljung BM, et al. Genetic alterations in ERBB2-amplified breast carcinomas. Clin Cancer Res 1999; 5: 4140–4145.
Tubbs R, Pettay J, Roche P, Stoler MH, Jenkins RB, Grogan TM . Discrepancies in clinical laboratory testing of eligibility for trastuzumab therapy: apparent immunohistochemical false-positives do not get the message. J Clin Oncol 2001; 19: 2714–2721.
Tirkkonen M, Tanner M, Karhu R, Kallioniemi A, Isola J, Kallioniemi OP . Molecular cytogenetics of primary breast cancer by CGH. Genes Chromosomes Cancer 1998; 21: 177–184.
Gramlich TL, Cohen C, Fritsch C, DeRose PB, Gansler T . Evaluation of c-erbB2 amplification in breast carcinoma by different polymerase chain reaction. Am J Clin Pathol 1994; 101: 493–499.
Persons DL, Borelli KA, Hsu PH . Quantitation of HER-2/neu and c-myc gene amplification in breast carcinoma using fluorescence in situ hybridization. Mod Pathol 1997; 10: 720–727.
Benz CC . The molecular biology of HER-2. Presented at 21st San Antonio Breast Cancer Symposium, San Antonio, Texas, December 12–15, 1998.
Roche PC, Ingle JN . Increased HER2 with U.S. Food and Drug Administration-approved antibody. J Clin Oncol 1999; 17: 434–435.
Slamon DJ, Leyland-Jones B, Shack S . Addition of Herceptin (humanized anti-HER2 antibody) to first line chemotherapy for HER2 overexpressing metastatic breast cancer (HER2+/MBC) markedly increases anticancer activity: a randomized multinational controlled phase II trial. Proc Am Soc Clin Oncol 1998; 17: 98a.
Maia DM . Immunohistochemistry assays for HER2 overexpression. J Clin Oncol 1999; 17: 434.
United States Food and Drug Administration, Department of Health and Human Services. Oncology Drugs Advisory Committee, 58th Meeting, September 2, 1998, P.M. session. Available at: www.fda.gov/ohrms/dockets/ac/98/transcrpt/3443t2.rtf.
Check W . More than one way to look for HER2. CAP Today 1999; 13: 48–52.
U S PMA. DAKO HercepTest [package insert]. Carpinteria, CA: DAKO; 1998.
Tripathy D . Paclitaxel plus Trastuzumab in HER2 overexpressing and non-overexpressing metastatic breast cancer. 22nd Annual San Antonio Breast Cancer Symposium, San Antonio, Texas; 1999.
Kumamoto H, Sadano H, Takahiro T, Suzuki T, Moriya T, Ichinohasama R . Chromogenic in situ hybridization analysis of HER-2/neu status in breast carcinoma: application in screening of patients for trastuzumab (Herceptin) therapy. Pathol Int 2001; 51: 579–584.
Acknowledgements
The authors thank Dr. Jorma Isola, Dr. Jonathan A. Fletcher, and Dr. David Weinstein for the valuable discussion and for reviewing the manuscript. The authors thank Bill Arnold for preparation of images used in this paper.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhao, J., Wu, R., Au, A. et al. Determination of HER2 Gene Amplification by Chromogenic In Situ Hybridization (CISH) in Archival Breast Carcinoma. Mod Pathol 15, 657–665 (2002). https://doi.org/10.1038/modpathol.3880582
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.3880582
Keywords
This article is cited by
-
Nucleic Acids Analysis
Science China Chemistry (2021)
-
The clinical and biological significance of HER2 over-expression in breast ductal carcinoma in situ: a large study from a single institution
British Journal of Cancer (2019)
-
Identification of aldolase A as a potential diagnostic biomarker for colorectal cancer based on proteomic analysis using formalin-fixed paraffin-embedded tissue
Tumor Biology (2016)
-
HER2 screening data from ToGA: targeting HER2 in gastric and gastroesophageal junction cancer
Gastric Cancer (2015)
-
A new bright-field dual-colour chromogenic and silver in situ hybridization method for the detection of FGFR1 gene copy number status
Virchows Archiv (2014)