Abstract
Forty-nine cases of synovial sarcoma were evaluated for mutation of the p53 gene, amplification of the MDM2 gene and mutation of the H-ras gene, and for the relation of these factors to overall survival and clinicopathologic parameters. All investigations were carried out on formalin-fixed paraffin-embedded materials. Furthermore, we evaluated the expression of p53 protein, MDM2, and p21WAF1/CIP1 immunohistochemically in these cases, together with an assessment of proliferative activities using monoclonal antibody MIB-1. Nine of the 49 cases (18.4%) had p53 gene alteration detected by polymerase chain reaction-single-strand conformation polymorphism (PCR-SSCP) and direct sequencing. Eleven cases (24%) showed nuclear accumulation of p53 protein in more than 10% of the tumor cells. Among them, only three cases contained gene mutations. There was no correlation between p53 nuclear accumulation and p53 gene alteration. MDM2 gene amplification, as shown by differential PCR, was observed in 19 out of 47 cases (40%). Nineteen out of 49 cases (38.8%) showed immunoreactivity for MDM2. MDM2 gene amplification and the expression of MDM2 protein showed a significant positive relationship (P = 0.0004). Moreover, MDM2 immunoreaction was significantly correlated with nuclear accumulation of p53 protein (P = 0.023). Positive immunoreaction for p21WAF1/CIP1 was observed in 21 out of 48 cases (43.8%). p21WAF1/CIP1 expression was correlated with p53 protein expression. H-ras gene mutations were seen in only three cases (6.1%). All mutations were in codon 12 (one GGC-to-AGC [Gly-to-Ser] mutation and two GGC-to-GAC [Gly-to-Ap] mutations). The gene alteration of p53, MDM2, and H-ras did not affect the patients’ prognosis. Although the cases with positive immunoreaction for p53 tended to have a worse prognosis, the difference was not statistically significant (P = 0.13). No correlation was observed between MIB-1 LI and the immunohistochemical expression of p53, MDM2, and p21WAF1/CIP1 or the mutation status of p53 and H-ras. On the other hand, high MIB-1 LI (more than 10) significantly correlated with poor prognosis (P < 0.0001). Our results suggest that p53 gene mutation does not appear to be a major prognostic factor and H-ras mutations are infrequent in synovial sarcoma.
Similar content being viewed by others
INTRODUCTION
Mutations of the p53 gene are the most common alteration found in human malignant tumors and alterations of p53 have been suggested as having a potential role to play in tumor progression. The MDM2 gene was originally isolated from mouse double minute chromosomes, the cytogenetic hallmark of gene amplification (1). Its protein products bind to p53 protein, forming a complex that can inhibit the p53 growth suppressive function by blocking the transactivation domain of p53 (2, 3). On the other hand, the coexpression of both p53 and MDM2 protein has been demonstrated as being an adverse prognostic factor in studies of several types of soft tissue sarcomas (4, 5).
p21, which is known as WAF1 (6) and CIP1 (7), and which can be transactivated by the tumor suppressor gene p53 in response to DNA damage (6), is a potent inhibitor of all cyclin-dependent kinases. Alternative, p53-independent mechanisms of p21WAF1/CIP1 induction have also been described (8). Although, p53 and p53 protein together with the p53-related proteins MDM2 and p21WAF1/CIP1 have been analyzed in several kinds of sarcoma (9, 10), there have been a few investigations in a series of synovial sarcoma (11, 12, 13, 14).
The ras gene family (K-ras, H-ras, and H-ras) has been reported to be associated with human neoplasms (15). Frequent ras mutations have been reported in a number of human cancers, including adenocarcinoma of the pancreas (90%), colon (50%), thyroid (50%), and lung (30%) (16). The H-ras gene is commonly activated in human urinary tract tumors (17). As for sarcomas, H-ras codon 12 mutations have been reported in rhabdomyosarcoma and malignant fibrous histiocytoma (18, 19). However, data of H-ras mutation in synovial sarcoma is currently unavailable.
The aim of this study was to define the frequency of p53, MDM2, and H-ras gene alteration in a rather large series of synovial sarcoma and to investigate their possible association with clinicopathologic features and proliferative activities.
MATERIALS AND METHODS
Case Materials
Forty-nine formalin-fixed, paraffin-embedded synovial sarcoma specimens were available for both immunohistochemical and molecular studies. The diagnosis of all cases was based on light microscopic examination with a hematoxylin-eosin (HE) stain and, when necessary, immunoperoxidase procedures using the streptavidin-biotin peroxidase (SAB) method were carried out. In 21 cases of the current series, reverse transcription polymerase reaction (RT-PCR) for the detection of SYT-SSX transcripts which were specific for synovial sarcoma were performed using concordant frozen material (20). Survival data were available for 46 cases. Follow-up ranged from 1 to 233 mo (mean, 46.5 mo).
In order to assess the correlation between clinicopathologic factors and the results of both immunohistochemical and molecular analysis, sex, age, anatomical site, tumor depth, histologic subtype, the presence of rhabdoid cells, mitotic rate, nuclear grade, tumor necrosis, the number of mast cells, tumor size, and tumor stage according to the American Joint Committee (AJC) were analyzed. The influence of these factors on prognosis has been previously reported (21).
Polymerase Chain Reaction-Single-Strand Conformation Polymorphism (PCR-SSCP) for p53
Genomic DNA was purified from paraffin-embedded tissues of 49 synovial sarcomas, using standard proteinase K digestion and phenol/chloroform extraction.
Obtained DNA was subjected in a total volume of 5 μL containing 10 mm Tris-HCl (pH 8.3), 50 mm KCl, 2.0 mm MgCl2, 25 mm of dNTP, 0.25 U Taq DNA polymerase, and 0.2 μm of each of the primers. Mutations of the p53 gene were examined from exons 5 to 9. The sequences of the primers are summarized in Table 1. PCR was carried out using a programmable thermal cycler (PTC-100TM: MJ Research, Inc., Watertown, MA) for 40 cycles after the first denaturation at 95° C for 1 min. Each cycle consisted of denaturation at 95° C for 1 min, at 66° C for 1 min, and at 72° C for 2 min. Obtained products were electrophoresed through 2.0% agarose gel with ethidium bromide at 100V for 50 min. The DNA band was purified using a SUPREC tube (TAKARA Biomedicals, Japan) and the resulting products were reamplified for 15 cycles. Three μL of the reamplified products were diluted 1:1 in loading buffer (94% formamide, 10 mg bromphenol blue, and 0.05% xylene cyanol), heated at 96° C for 5 min, snap frozen on dry ice, and then a total of 6 μL of samples was loaded onto the gel containing 12.5% acrylamide (GenePhorTM, Amersham Pharmacia Biotech Uppsala, Sweden). SSCP was performed using a DNA fragment analyzer (GenePhorTM, Amersham Pharmacia Biotech Uppsala, Sweden) at 600V, 25mA, 15W, 5° C, for 120 min, and then visualized by a DNA Silver Staining Kit (GenePhorTM, Amersham Pharmacia Biotech).
Differential PCR for MDM2
The differential PCR method to detect amplification of MDM2 in this study was a modification of the method of Reid et al. (22). DNA samples were added to a PCR mix with a total volume of 25 μL containing 10 mm Tris-HCl (pH 8.3), 50 mm KCl, 1.5 mm MgCl2, 0.25 mm dNTP, 0.5U Taq DNA polymerase, and 0.5 μm of each of the primers. The sequences of the primers used were as follows: MDM2, 5′-GGTTGGATCAGGATTCAGTT-3′ (sense) and 5′-GAGTTCTTGTCCTTCTTCAC-3′ (antisense); PAH as internal control, 5′-ATGCCACTGAGAACTCTTT-3′ (sense) and 5′-GAGTTCTTGTCCTTCTTCAC-3′ (antisense). Samples were placed in a thermal cycler (PTC-100TM: MJ Research), heated to 95° C for 1 min, then cycled for 1 min each at 94° C, 50° C, and 70° C for 30 cycles. Each set of samples included one tube with no added DNA template, which served as a negative control. As for positive control, DNA of the SA-1 cell line (American Type Cell Collection, Rockville, MD), which is known to show seven-fold amplification of the MDM2 gene by the radio-labeled differential PCR method (22), was used. After amplification, 10 μL of each reaction were analyzed by electrophoresis on a 3% agarose gel, stained with ethidium bromide, and the intensities of the DNA products were quantified using a National Institutes of Health (NIH) Image Ver 1.56. The level of MDM2 amplification was determined by comparing the ratio of the intensities of the MDM2 and PAH PCR products for each of the samples with positive SA-1 cells (seven-fold). Samples showing more than two-fold amplification were judged as positive.
Polymerase Chain Reaction-Restriction Fragment Length Polymorphism (PCR-RFLP) for H-ras
We used the PCR-RFLP procedure to detect H-ras mutations at codons 12 and 13 with strategy primers as reported (23). DNA sequences containing codons 12 and 13 of the H-ras gene were amplified using the primers 5′-GGAGACCCTGTAGGAGGACCC-3′ and 5′-TCTATAGTGGGGTCGTATTCGTCC-3′ for 35 cycles (95° C for 1 min, 59° C for 1 min, and 72° C for 2 min). Then 5 μL of the 100 × diluted amplified products were reamplified by means of a nested PCR, using mutant primers 5′-TGAGGAGCGATGACGGAAT-3′ and 5′-ATGGTCAGCGCACTCTTGCCCTC-3′ for 30 cycles (92° C for 15 s, 55° C for 15 s, and 72° C for 30 s). Codon 12 mutations were detected thanks to a naturally occurring HpaII site (CCGG) that is lost when the mutation occurs. HpaII digests the 71-bp amplified fragment into two fragments (41-bp and 30-bp), thus revealing the presence of the normal allele, while the mutant allele remains within the undigested 71-bp fragment. This method allows detection of a mutation of the second and third positions of codon 11 and the first and second positions of codon 12. To detect codon 13 mutation, nested primers were used to create a new restriction site for HphI (GGTGA) by changing a T for an A in the second position of codon 14, thereby allowing detection of any mutation of codon 13 of the H-ras gene. Hph I digests the same 71-bp amplified fragment into two fragments (58-bp and 13-bp), thus revealing the presence of the normal allele, while the mutant allele remains within the undigested 71-bp fragment. The DNA bands were analyzed by 3% agarose gel electrophoresis, stained with ethidium bromide and photographed.
DNA Sequencing
Aberrantly migrating bands of p53 were excised from the SSCP gel, and reamplified for 25 cycles under the same conditions. As for H-ras, the digested products from agarose gels were amplified by the same primers used for nested-PCR. The products after reamplification were further purified by microcon 100 (AMICON, Millipore, Bedford, MA). After the purification, direct sequencing was carried out by the dideoxy chain termination methods using a Perkin Elmer ABI Prism 310 sequence analyzer (Applied Biosystems, Foster City, CA). The primers used for direct sequences were the sense and anti-sense primers used for the nested PCR in H-ras and PCR-SSCP in p53.
Immunohistochemistry
The following monoclonal antibodies were used: anti-Ki-67 antibody MIB-1 (1:100: Immunotech, Marseille, France), anti-p53 (Pab1801, 1:100, Oncogene Science, New York, NY), anti-MDM2 (1:40, Oncogene Research Products, Cambridge, MA), and anti-WAF1 (1:100, Oncogene Research Products). Five-μm thick histologic sections of 10% formalin-fixed, paraffin-embedded materials were cut, mounted on glass slides coated by 3-aminopropyltriethoxysilane, and air-dried overnight at room temperature. The sections were deparaffinized in xylene and rehydrated in ethanol, and endogenous peroxidase was blocked by methanol containing 3% hydrogen peroxidase for 15 min. For staining the above antibodies, sections were pretreated with citrate buffer (0.01mol/L citric acid: pH 6.0) four times, each for 5 min at 100° C in a microwave oven. The sections were incubated with primary antibodies at 4° C overnight, followed by staining with a streptavidin-biotin-peroxidase kit (Nichirei, Tokyo, Japan). The sections were then finally reacted in a 3,3′-diaminobenzidine, peroxytrichloride substrate solution and counterstained with methyl green. As for p53, MDM2, and WAF1, staining of more than 10% nuclei was needed for positive cases. The MIB-1 labeling index (MIB-1 LI) was determined by counting the positively stained nuclei in at least 500 tumor cells.
Statistical Analysis
Fisher’s exact test was used to evaluate the association between two dichotomous variables. Concerning the histologic subtype, one poorly differentiated subtype was excluded from the statistical analysis because the number of samples was too small to analyze. The difference in the MIB-1 LI between two groups was estimated by unpooled t test. Survival curves were calculated by the Kaplan-Meier method, and survival differences were evaluated by the log-rank test. A P value of less than 0.05 was considered statistically significant.
RESULTS
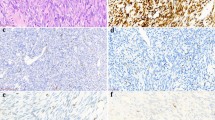
There were 20 males and 29 female patients, ranging in age from 7 to 65 years (mean, 36.5 years). Thirty-one tumors had arisen within the proximal extremities or trunk, whereas 18 had occurred within the distal extremities. Forty-three tumors were deeply situated, whereas four were superficially situated. The depth of two tumors was unknown. Thirty-one tumors were 5 cm or larger. The size of six tumors was unknown. Histologically, 40 tumors were categorized as monophasic fibrous tumors, eight as biphasic tumors, and one as a poorly differentiated tumor (Fig. 1). The mitotic rate was classified as high in 15 cases with more than 15/10HPF. Eleven tumors demonstrated nuclear grade III, whereas 38 demonstrated grade I or II. Tumor necrosis was extensive in 10 cases, with more than 50% tumor necrosis on the examined slides. Rhabdoid cells with abundant glassy eosinophilic cytoplasm and eccentric nuclei were present in six tumors. The frequent existence of mast cells with more than 20/10HPF was recognized in 13 tumors. Twenty-two tumors were stage I or II, whereas 18 were stage III or IV.
A, monophasic fibrous type synovial sarcoma composed of spindle cells, arranged in a fascicle. B, biphasic type synovial sarcoma with glandular structure lined by epithelioid cuboidal cells. C, poorly differentiated type showing a proliferation of predominantly rounded or polygonal cells (hematoxylin and eosin stain; original magnification, 300 ×).
p53 Mutations in Exons 5–9
Nine of the 49 cases (18.4%) had p53 gene abnormalities detected by molecular biological analysis (Fig. 2, A–D). There were 11 mutational sites, one being nonsense mutation (Case 11), two being silent mutations (Cases 16 and 41), and 8 being missense mutations (Table 2). One tumor (Case 52) contained three mutations at codons 141, 159, and 173. As for the type of base change, there were 10 sites of transition and one site of transversion. No positive correlation was observed between p53 mutation status and clinicopathologic parameters (Table 3) or prognosis.
PCR-SSCP analysis at exon 7 (A) and exon 6 (B). Abnormally shifted bands are recognized at S26 (A) and S36 (B), respectively. HG: Human genomic DNA as a positive normal control. Direct DNA sequencing of exon 7 (C) in Case S26 and of exon 6 (D) in Case S36. A to G transition was detected at codon 235 substituting Asp for Asn (C). A to T transversion was seen at codon 195 substituting Phe to Ile (D).
p53 Immunoreactivity
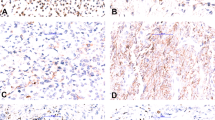
Eleven cases (22.4%) revealed nuclear accumulation of p53 protein in more than 10% of tumor cells (Fig. 3). Among them, three cases contained five p53 mutations comprising one nonsense mutation and four missense mutations. There was no correlation between p53 immunoreactivity and p53 mutation status (Table 4). Although the cases with immunoreaction with p53 protein tended to have worse prognosis, the difference was not statistically significant (Fig. 4; P = 0.13). There was no relationship between p53 immunoreaction and clinicopathologic parameters.
MDM2 Immunoreactivity and Amplification
Nineteen out of the 49 cases (38.8%) showed immunoreaction for MDM2 (Fig. 5). MDM2 expression was correlated with a young age of less than 20 years, proximal location of the tumor, and biphasic subtype (Table 3). No correlation was observed between MDM2 immunoreaction and prognosis. Differential PCR for MDM2 was available in 47 cases. Nineteen cases (40%) showed MDM2 amplification (Fig. 6) and amplification of the MDM2 gene had a significantly positive correlation with MDM2 immunoreactivity (Table 5; P = 0.0004). MDM2 amplification had no correlation with clinicopathologic parameters or prognosis. MDM2 immunoreaction also had a significant relationship with nuclear accumulation of p53 protein (Table 4; P = 0.023), but not with p53 mutation status. Coexpression of p53 and MDM2 was observed in eight cases (16.3%); however, coexpression of both proteins did not affect overall survival (P = 0.413).
Differential PCR for 10 synovial sarcoma cases. MDM2 is amplified in Cases S4, S5, S6, S8, and S10. In these cases, the intensities of MDM2 are more than two-fold those of the concordant PAH gene. For quantitation, the intensity of each band was analyzed by NIH Imaging. P: Positive control. PCR product of the reaction with DNA from the SA-1 cell line, which is known to amplify the MDM2 gene 7-fold. N: Negative control. PCR product from the reaction without template DNA.
Expression of p21WAF1/CIP1
Immunohistochemical study for p21WAF1/CIP1 could be carried out in 48 cases. Twenty-one out of these 48 cases (43.8%) revealed positive immunoreaction for p21WAF1/CIP1 (Fig. 7). p21WAF1/CIP1 expression was positively correlated with nuclear accumulation of p53 protein and p53 mutation (Table 4, P = 0.001 and Table 6, P = 0.022, respectively). Co-assessment of p21WAF1/CIP1 and p53 subgroups revealed that p21WAF1+/p53- patients seemed to have a better survival compared with other groups (Fig. 8); however, the difference was not statistically significant (P = 0.209).
H-ras Mutation Status
Overall H-ras mutations were detected in three out of the 49 cases (6.1%) (Fig. 9). Table 7 summarizes the data of these three cases. There were three mutations in codon 12 (one GGC-to- AGC [Gly-to-Ser] and two GGC-to-GAC [Gly-to-Asp]). Two patients died of disease at 11 mo and 40 mo, whereas one patient remains alive at 32 mo without any evidence of disease. There was no relationship between H-ras mutation status and clinicopathologic parameters.
MIB-1 LI
MIB-1 LI ranged from 2.1 to 36.7 (mean, 12.9). Different cut-off points of MIB-1 LI (10, 15, 20, and 25) were evaluated. All cut-off values showed significant influence on the patient’s survival (P < 0.0001). High MIB-1 LI of more than 10 was also significantly correlated with poor survival (log-rank test: P < 0.0001) (Fig. 10). No correlation was observed between MIB-1 LI and the immunohistochemical expression of p53, MDM2, and p21WAF1/CIP1 or the mutation status of p53 and H-ras (Table 8).
DISCUSSION
More than 90% of synovial sarcomas are characterized by the chromosomal translocation t(X;18)(p11;q11) as the sole cytogenetic abnormality (24). RT-PCR method was employed to detect the SYT-SSX fusion transcript. Kawai et al. (25) found that the patients with SYT -SSX1 transcripts had a worse prognosis, compared with SYT-SSX2 patients group. However, Oda et al. (20) and Tsuji et al. (26) could not demonstrate any relationship between fusion gene phenotype and prognosis. In synovial sarcoma, other molecular prognostic factors also controversial.
p53 mutation status and its nuclear accumulation have been extensively evaluated in soft tissue sarcomas. Most of these reports have deal with a mixture of several kinds of soft tissue sarcomas (4, 5, 27, 28, 29), although some investigations have studied only a single type of sarcoma such as dermatofibrosarcoma protuberans (10), malignant fibrous histiocytoma (22), leiomyosarcoma (30), or liposarcoma (9, 31). With regard to synovial sarcoma, a few authors have presented data regarding p53 expression and mutation. Kawai et al. (28) revealed the nuclear accumulation of p53 protein in three out of six synovial sarcomas in their series of 96 soft tissue sarcomas. Cordon-Cardo et al. (4) examined the immunoreactivity for p53 and MDM2 in 211 cases of soft tissue sarcoma including 13 synovial sarcomas, however, they did not report the number of p53-positive cases among the synovial sarcoma cases. Pollock et al. (32) presented a case of synovial sarcoma with p53 point mutation in codon 135. Recently, Antonesque et al. (11) and Dei Tos et al. (12) demonstrated the expression of p53 protein in eight of 49 synovial sarcomas (16%) and in 21 out of 41 cases (51%), respectively. In addition, Dei Tos et al. (12) and Schneider-Stock et al. (13) detected p53 mutations in four of 20 cases (20%) and in two of 34 cases (5.9%), respectively, with the PCR-SSCP method. In our series, nuclear accumulation of p53 was observed in 11 out of the 49 cases (24%), whereas p53 mutation was detected in nine of the 49 cases (18.4%).
The prognostic value of p53 nuclear accumulation or its mutation in soft tissue sarcoma is controversial. Kawai et al. (28) showed that nuclear immunoreaction of p53 protein was correlated with poor prognosis in the mixture of soft tissue sarcomas. Antonescue et al. (11) and Schneider-Stock et al. (13) reported that p53 overexpression can be a prognostic factor in synovial sarcoma. In this study, the cases with immunoreactivity for p53 protein seemed to have a worse prognosis compared with negative cases; however, the difference was not statistically significant. Skytting et al. (14) failed to reveal p53 immunoreactivity as prognostic factor. Taubert et al. (29) showed that patients with soft tissue sarcoma, where there was a non-frame-shift mutation of p53 gene had a significantly worse prognosis, while Konomoto et al. (30) demonstrated that an abnormal p53 gene was a useful prognostic factor in deep type leiomyosarcoma. The only type of p53 mutation in this study was non-frame-shift mutation, however, no correlation was observed between prognosis and p53 mutation status.
Although the immunohistochemical expression of p53 protein was not correlated with p53 gene mutation, it had a significant relationship with MDM2 expression in this study. This finding is similar to that noted with retroperitoneal well- and de-differentiated liposarcoma (31), and supports the hypothesis that the sequestration of p53 by MDM2 protein, as observed in cell lines, may represent an alternative mechanism leading to the loss of p53 function. In addition to direct MDM2-p53 protein interactions via complexing, p53 controls the transcriptional activity of MDM2, resulting in an autoregulative balance feedback loop (33). Disturbance of the regulated balance between p53 and MDM2 can result in uncontrolled cell growth and subsequent tumorigenesis. Cordon-Cardo et al. (4) and Würl et al. (5) showed that coincident MDM2/p53 overexpression is of high prognostic relevance in soft tissue sarcoma. In the current study, the coexpression of these two proteins did not affect the prognosis of the patients.
p21 protein product is believed to block cyclin/cyclin-dependent kinase (CDK) complex activity and thus prevents the passage of cycling cells from the G1 to the S phase in a p53-dependent or p53-independent manner (6, 7, 8). In this study, p21WAF1/CIP1 expression was positively correlated with both the expression of p53 protein and p53 mutation, despite the discrepancies between p53 protein expression and mutation status. The reason for this phenomenon is unknown; however, p21WAF1/CIP1 expression had no correlation with proliferating activity or patients’ prognosis. Recently, McClelland et al. (34) reported the significance of p21 expression in a population of endocrine therapy-treated advanced breast cancers. They demonstrated that the subgroup of p21+/p53- patients has good survival characteristics. In our series of synovial sarcoma, this subgroup tended to have better prognosis compared with other subgroups, although there was no statistical significance. Pindzola et al. (35) analyzed the immunohistochemical expression of p21WAF1/CIP1 in 66 soft tissue sarcomas. They could not find any correlation between Ki-67 and p21WAF1/CIP1 or p53 immunostaining, as was also the case in our study. They reported that all their six cases of synovial sarcoma expressed p21WAF1/CIP1. On the other hand, in our series, 21 out of 48 cases (43.8%) showed immunoreaction for p21WAF1/CIP1; however, our cut-off value of p21WAF1/CIP1 was different from that in their study.
Within the ras family gene, H-ras gene mutation was frequently reported in urinary tract tumors (17). Concerning soft tissue sarcomas, H-ras mutations were detected in one out of five cases of rhabdomyosarcoma (embryonal type: G-to-T mutation in the second position of codon 12) (18) and two out of six cases of malignant fibrous histiocytoma (MFH) (G-to-A mutations in the second position of codon 12) (18), using snap-frozen materials combined with PCR and direct sequencing. Bohle et al. (19) searched for point mutations of the ras genes including H-ras-1, K-ras-2, and N-ras in 32 cases of MFH, employing formalin-fixed, paraffin-embedded material with nested PCR and direct sequencing. They detected nine mutations (28%) in two of 12 pleomorphic type, four of nine myxoid type, and one of six xanthomatous type cases. All the mutations were H-ras codon 12 mutations with G-to-T mutation in the second position. Yoo and Robinson (36) reported H-ras mutation in 6 MFHs and one leiomyosarcoma among 45 Korean soft tissue sarcoma patients, but only in one rhabdomyosarcoma among 50 US soft tissue sarcoma patients. There have been no previous investigations evaluating H-ras mutation in synovial sarcoma. In this study, H-ras mutations were detected in three out of 49 synovial sarcomas (6.1%) and this mutation was less frequent compared with MFH. All three cases showed the mutation in codon 12 (one G-to-A mutation in the first position and two G-to-A mutations in the second position). There was no correlation between H-ras mutation status and clinicopathologic parameters or prognosis.
A few authors reported that proliferating activities evaluated by proliferating cell nuclear antigen (PCNA) (37) and Ki-67 (14) are closely related with patients’ prognosis in synovial sarcoma. Skytting et al. (14) demonstrated a significant difference in the metastasis-free survival rate for patients with a high MIB-1 index (cut-off value: 10%) versus patients with a low index. We evaluated four cut-off values of MIB-1 LI (10, 15, 20, and 25) and found all cut-off points correlated with the patients’ survival. Consequently, high MIB-1 LI also significantly correlated with poor prognosis in our series as shown by Skytting et al. (14).
In conclusion, p53 gene mutations are relatively rare and do not affect the patients’ prognosis and H-ras mutation is infrequent in synovial sarcoma. In addition, it is possible that MDM2-mediated inactivation of p53 may occur in some cases.
References
Farkharzadeh SS, Trusko SP, George DL . Tumorigenic potential associated with enhanced expression of a gene that is amplified in mouse tumor cell line. EMBO J 1991; 10: 1565–1569.
Olson DC, Marechal V, Momand J, Chen J, Romocki C, Levine AJ . Identification and characterization of multiple mdm-2 proteins and mdm-2-p53 protein complex. Oncogene 1993; 8: 2353–2360.
Picksley SM, Vojtesek B, Sparks A, Lane DP . Immunochemical analysis of the interaction of p53 with MDM2; – fine mapping of the MDM2 binding site on p53 using synthetic peptides. Oncogene 1994; 9: 2523–2529.
Cordon-Cardo C, Latres E, Drobnjak M, Oliva MR, Pollack D, Woodruff JM, et al. Molecular abnormalities of mdm2 and p53 genes in adult soft tissue sarcomas. Cancer Res 1994; 54: 794–799.
Würl P, Meye A, Schmidt H, Lautenschläger C, Kalthoff H, Rath F-W, et al. High prognostic significance of Mdm2/p53 co-overexpression in soft tissue sarcomas of the extremities. Oncogene 1998; 16: 1183–1185.
El-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, et al. WAF1, a potential mediator of p53 tumor suppression. Cell 1993; 75: 817–825.
Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ . The p21 cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 1993; 75: 805–816.
Parker SB, Eichele G, Zhang P, Rawls A, Sands AT, Bradley A, et al. p53-independent expression of p21 Cip1 in muscle and other terminally differentiating cells. Science 1995; 267: 1024–1027.
Dei Tos AP, Doglioni C, Piccinin S, Maestro R, Mentzel T, Barbareschi M, et al. Molecular abnormalities of the p53 pathway in dedifferentiated liposarcoma. J Pathol 1997; 181: 8–13.
Hisaoka M, Okanoto S, Morimitsu Y, Tsuji S, Hashimoto H . Dermatofibrosarcoma protuberans with fibrosarcomatous areas. Molecular abnormalities of the p53 pathway in fibrosarcomatous transformation of dermatofibrosarcoma protuberans. Virchows Arch 1998; 433: 323–329.
Antonescu CR, Leung DH, Dudas M, Woodruff JM, Cordon-Cardo C . Cell cycle dependent molecular expression in localized synovial sarcoma. A multifactorial study with prognostic implications [abstract]. Mod Pathol 1999; 12: 9A.
Dei Tos AP, Doglioni C, Muffato R, Maestro R, Furlanetto A, Sciot R, et al. Synovial sarcoma exhibits deregulation of the G1-S cell cycle checkpoint and of the apoptotic pathway [abstract]. Mod Pathol 1999; 12: 10A.
Schneider-Stock R, Onnasch D, Haeckel C, Mellin W, Franke D-S, Roessner A . Prognostic significance of p53 gene mutations and p53 protein expression in synovial sarcomas. Virchow Archiv 1999; 435: 407–412.
Skytting BT, Bauer HC, Perfekt R, Nilsson G, Larsson O . Ki-67 is strongly prognostic in synovial sarcoma: analysis based on 86 patients from Scandinavian Sarcoma Group Register. Br J Cancer 1999; 80: 1809–1814.
Barbacid M . ras genes. Annu Rev Biochem 1987; 56: 779–827.
Bos JL . ras oncogenes in human cancer: a review. Cancer Res 1989; 49: 4682–4689.
Fujita J, Yoshida O, Yuasa Y, Rhim JS Hatanaka M, Aaronson SA . Ha-ras oncogenes are activated by somatic alterations in human urinary tract tumors. Nature 1984; 309: 464–466.
Wilke W, Maillet M, Robinson R . H-ras-1 point mutations in soft tissue sarcomas. Mod Pathol 1993; 6: 129–132.
Bohle RM, Brettreich S, Repp R, Borkhardt A, Kosmehl H, Altmannsberger HM . Single somatic ras gene point mutation in soft tissue malignant fibrous histiocytoma. Am J Pathol 1996; 148: 731–738.
Oda Y, Sakamoto A, Saito T, Kinukawa N, Iwamoto Y, Tsuneyoshi M . Expression of hepatocyte growth factor (HGF)/scatter factor and its receptor c-MET correlates with poor prognosis in synovial sarcoma. Hum Pathol 2000; 31: 185–192.
Oda Y, Hashimoto H, Tsuneyoshi M, Takeshita S . Survivals in synovial sarcoma. A multivariate study of prognostic factors with special emphasis on the comparison between early death and long-term survivals. Am J Surg Pathol 1993; 17: 35–44.
Reid AH, Tsai MM, Venzon DJ, Wright CF, Lack EE, O’Leary TJ . MDM2 amplification, p53 mutation, and accumulation of the p53 gene product in malignant fibrous histiocytoma. Diag Mol Pathol 1996; 5: 65–73.
Capella G, Matias-Guiu X, Ampudia X, de Leiva A, Perucho M, Prat J . Ras oncogene mutations in thyroid tumors. Diag Mol Pathol 1996; 5: 45–52.
Sreekantaiah C, Ladanyi M, Rodriguez E, Chaganti RSK . Chromosomal aberrations in soft tissue tumors. Relevance to diagnosis, classification, and molecular mechanisms. Am J Pathol 1994; 144: 1121–1124.
Kawai A, Woodruff J, Healey JH, Brennan F, Antonescu CR, Ladanyi M . SYT-SSX gene fusion as a determinant of morphology and prognosis in synovial sarcoma. N Engl J Med 1998; 338: 153–160.
Tsuji S, Hisaoka M, Morimitsu Y, Hashimoto H, Shimajiri S, Komiya S, et al. Detection of SYT-SSX fusion transcripts in synovial sarcoma by reverse transcription-polymerase chain reaction using archival paraffin-embedded tissues. Am J Pathol 1998; 153: 1807–1812.
Toguchida J, Yamaguchi T, Ritchie B, Beauchamp RL, Dayton SH, Herrera GE, et al. Mutation spectrum of the p53 gene in bone and soft tissue sarcomas. Cancer Res 1992; 52: 6194–6199.
Kawai A, Noguchi M, Beppu Y, Yokoyama R, Mukai K, Hirohashi S, et al. Nuclear immunoreaction of p53 protein in soft tissue sarcomas. A possible prognostic factor. Cancer 1994; 73: 2499–2505.
Taubert H, Meye A, Würl P . Prognosis is correlated with p53 mutation type for soft tissue sarcoma patients. Cancer Res 1996; 56: 4134–4136.
Konomoto T, Fukuda T, Hayashi K, Kumazawa J, Tsuneyoshi M . Leiomyosarcoma in soft tissue: examination of p53 status and cell proliferating factors in different locations. Hum Pathol 1998; 29: 74–81.
Pilotti S, Torre GD, Lavarino C, Palma SD, Sozzi G, Minoletti F, et al. Distinct mdm2/p53 expression pattern in liposarcoma subgroups: implications for different pathogenetic mechanisms. J Pathol 1997; 181: 14–24.
Pollock RE, Lang A, Luo J, El-Naggar AK, Yu D . Soft tissue sarcoma metastasis from clonal expansion of p53 mutated tumor cells. Oncogene 1996; 12: 2035–2039.
Oliner JD, Pietenpol JA, Thiagalingam S, Gyuris J, Kinzler KW, Vogelstein B . MDM2 conceals the activation domain of tumor suppressor p53. Nature 1993; 362: 857–860.
McClelland RA, Gee JMW, O’Sullivan L, Barnes DM, Robertson JFR, Ellis IO, et al. p21WAF1 expression and endocrine response in breast cancer. J Pathol 1999; 188: 126–132.
Pindzola JA, Palazzo JP, Kovatich AJ, Tuma B, Nobel M . Expression of p21WAF1/CIP1 in soft tissue sarcomas: a comparative immunohistochemical study with p53 and Ki-67. Path Res Pract 1998; 194: 685–691.
Yoo J, Robinson RA . H-ras, and K-ras mutations in soft tissue sarcoma. Comparative studies of sarcomas from Korean and American patients. Cancer 1999; 86: 58–63.
Oda Y, Hashimoto H, Takeshita S, Tsuneyoshi M . The prognostic value of immunohistochemical staining for proliferating cell nuclear antigen (PCNA) in synovial sarcoma. Cancer 1993; 72: 478–485.
Acknowledgements
The English used in this manuscript was revised by Miss K. Miller (Royal English Language Center, Fukuoka, Japan).
This work was supported in part by a Grant-in-Aid for Cancer Research from the Fukuoka Cancer Society, Fukuoka, and a Grant-in-Aid for General Scientific Research from the Ministry of Education, Science, Sports, and Culture (09470052 and 12670167), Tokyo, Japan.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Oda, Y., Sakamoto, A., Satio, T. et al. Molecular Abnormalities of p53, MDM2, and H-ras in Synovial Sarcoma. Mod Pathol 13, 994–1004 (2000). https://doi.org/10.1038/modpathol.3880180
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.3880180
Keywords
This article is cited by
-
Upregulation of ERK-EGR1-heparanase axis by HDAC inhibitors provides targets for rational therapeutic intervention in synovial sarcoma
Journal of Experimental & Clinical Cancer Research (2021)
-
GBAF, a small BAF sub-complex with big implications: a systematic review
Epigenetics & Chromatin (2020)
-
Establishment and characterization of NCC-SS3-C1: a novel patient-derived cell line of synovial sarcoma
Human Cell (2020)
-
An observational study on the expression levels of MDM2 and MDMX proteins, and associated effects on P53 in a series of human liposarcomas
BMC Clinical Pathology (2013)
-
p53 protein overexpression in bone marrow biopsies from chronic lymphocytic leukaemia is associated with TP53 deletion and resistance to fludarabine
Journal of Hematopathology (2010)