Abstract
Gastrointestinal stromal tumor (GIST) is the most common mesenchymal tumor in the gastrointestinal tract. The diagnosis of GIST is based on histology together with a panel of immunohistochemical markers; the most important is KIT (CD117). A total of 434 cases of GISTs were confirmed by histology and immunohistochemistry, and incorporated into tissue microarrays. Validation of histological features as well as the prognostic value of two immunohistochemical biomarkers (p16 and L1) was assessed. High mitotic rate, large tumor size, nuclear atypia, and small bowel primary site were all validated as negative prognostic factors in GISTs. Expression of p16 was significantly correlated with unfavorable prognosis, whereas L1 expression was not.
Similar content being viewed by others
Main
Mesenchymal neoplasms arising in the digestive tract are rare compared with epithelial neoplasms. Several entities exist, among which gastrointestinal stromal tumor (GIST) is the most common.1, 2 The diagnosis of GIST is often suspected histologically, but such tumors can have a broad morphologic spectrum. A panel of immunohistochemical markers is used to aid in correct diagnosis. A total of 91 to 98% of GISTs stain positively for KIT (CD117),3, 4 which is a major factor in the initial identification of GIST and therefore is often the inclusion criterion into many reported series. Staining of GISTs for other standard laboratory immunomarkers is more variable, including CD34 (≈70%), smooth-muscle actin (≈35%), S-100 (≈10%) and desmin (≈5%).2, 5, 6 PKC theta is an immunohistochemical marker found in two studies to be expressed in GISTs, KIT-negative GISTs and schwannomas, but not in other mesenchymal tumors of the GI tract.7, 8 Prognostic factors in GISTs are controversial, but the most accepted pathologic features are mitotic rate and tumor size. Small tumors with low mitotic activity usually have a benign behavior, and increasing the number of mitoses and increasing tumor size have been shown to be statistically significant.9 An evidence-based approach for defining the risk of aggressive behaviour in GISTs, based on tumor size and mitotic count, has been presented in a consensus document from multiple scientists in the field.10 Other factors as anatomic location, cellular atypia and necrosis have been shown to be independent prognostic factors.
Tissue microarrays are useful for rapid testing of the diagnostic and prognostic utility of antibodies in large numbers of archival samples. The arrayed tissue cores are histologically guided samples from representative regions of standard formalin-fixed paraffin-embedded tumor blocks. Kononen et al11 first described this technique, and has since been used in many studies requiring the evaluation of large series. Tissue microarrays have repeatedly been shown to be an efficient method in assessing the sensitivity and specificity of antibodies used in determining the origin or cell type of human tumors.12, 13
p16 is a tumor suppressor protein that inhibits cell cycling by arresting cells in G1 before entry into S phase. Genetic alterations resulting in diminished p16 levels are common in human cancers.14 Loss of p16 protein correlated with high-risk GIST in a small study by Sabah et al15 and later by Schneider-Stock et al.13 In this second, larger study with 284 GIST patients, p16 loss was found in 50% of the cases. These patients had a worse prognosis than those with p16-positive tumors, with a 2.3-fold increased risk of dying of disease. However, expression (rather than loss) of p16 correlates with unfavorable prognosis in high-grade breast carcinomas16 and also in most high-grade cervical intraepithelial neoplasia (CIN) and cervical carcinomas of squamous, glandular and small cell types.17 This implies that p16 loss is not required for oncogenesis and other mechanisms to dysregulate cell-cycle control (such as overexpression of cyclin D or loss of RB) may instead be employed.
L1 is a cell-surface adhesion molecule that is a member of the immunoglobulin superfamily, with six amino-terminal immunoglobulin-like domains followed by three to five fibronectin type III domains, a single transmembrane segment and a highly conserved cytoplasmic tail.18 The L1 antigen is expressed in neural cells and is involved in a number of different cellular processes, including neuronal cell migration, myelination, axon growth and pathfinding.19, 20 In addition, L1 has a role in the development of the renal collecting duct system.21 Expression of L1 has been demonstrated on paraffin-embedded tissue and in cell lines in a wide variety of normal and neoplastic tissues.22 The presence of L1 correlates with progression and metastatic spread of melanomas,23 and is associated with poor prognosis in ovarian and endometrial carcinomas.24 Recently, Kaifi et al25 evaluated L1 as a potential diagnostic marker and prognostic factor in a study with 72 GISTs. They reported L1 to be expressed in 73% of the GISTs, and these patients trended toward reduced survival.
The purpose of this study was to validate the clinical and immunohistochemical prognostic markers on a large series of defined GISTs from Norway.
Materials and methods
Patients and Samples
The study material consisted of selected tissue blocks from the archives of pathology departments in Norway. Cases were selected by evaluating the records of the Cancer Registry of Norway for mesenchymal tumors and poorly differentiated carcinomas in the gastrointestinal tract over a period of 30 years (1973–2002). A total of 3672 reports were evaluated, and all reports with clear evidence that the origin of the tumor was not mesenchymal were disqualified (based on the results of immunohistochemical staining and other methods reported at the time of primary diagnosis). A total of 1192 cases of candidate mesenchymal tumors were so identified, and slides and blocks from all these cases requested. The material was located in all of the 20 pathology departments in Norway. At the largest departments, a local pathologist examined the cases and a single representative block was forwarded. From two hospitals we received no material, which constituted 92 cases. In 129 cases (11%) no blocks were found and in addition in 102 cases (10%) the blocks did not contain enough material for further investigations. New slides of the remaining 869 cases were made and stained with hematoxylin and eosin (H&E), and histology re-reviewed by the first author. A total of 187 cases with inconsistent histologic features were additionally evaluated by the last author, and 146 of these cases were excluded from the study because they were almost certainly not mesenchymal tumors (based on the H&E staining), but rather carcinomas or lymphomas. Sixteen cases underwent additional independent expert review and three cases were thereby further excluded. A total of 720 cases were evaluated as representing true mesenchymal tumors of the GI tract, but for making tissue microarrays an additional 69 cases did not contain enough material for the required duplicate core extractions (blocks containing only small bite or core biopsies). The 651 remaining cases from the Cancer Registry were assembled into five tissue microarrays. For all cases, data were collected on age, date of death (if applicable), surgical treatment, location of primary tumor, size of tumor and history of other malignancies. Overall survival and metastatic status at presentation were recorded at the time of evaluating the records. Eight patients were treated with STI-571 (Glivec®), as this cohort largely predates the use of this drug. The Regional Committee for Medical Research Ethics, Northern Norway approved the study.
Construction of Tissue Microarrays
A slide with representative, viable tumor was selected from each case, and coupled to the corresponding formalin-fixed paraffin-embedded block. Duplicate 0.6-mm cores were taken from representative tumor tissue and inserted into a recipient paraffin block to create a tissue microarray, using a Beecher Instruments Micro Tissue Arrayer.26 The completed recipient blocks were sectioned at 4 μm and transferred to silanized glass slides. A total of five recipient blocks were made.
Histological Evaluation
The whole-section slides that were used for selecting representative tissue microarray cores were used for histological evaluation. Mitotic figures were counted in 50 consecutive high-power fields. Additional parameters were recorded in all cases and included the presence of spindled and/or epithelioid cell morphology. Focal or diffuse atypia was included in the review and defined as previously described by Miettinen et al.4 Other parameters such as necrosis, ulceration, mucosal invasion and hemorrhage were also assessed. The tumors were classified according to the published consensus risk-group stratification system based on maximum tumor size and mitotic count per 50 high-power fields.10
Immunohistochemical Staining and Scoring
Sections from the arrays were stained with H&E to confirm the presence of a representative tumor in each core. Further sections were stained with a panel of antibodies using a Ventana (Tucson, AZ, USA) automated immunohistochemical stainer according to the manufacturer's guidelines.
The antibodies used were the following: c-KIT (polyclonal, dilution 1:200; Dako), smooth-muscle actin (clone 1A4, dilution 1:200; Dako), S100 (polyclonal, dilution 1:2000; Dr A Marks, University of Toronto), CD34 (QB End/10, dilution 1:25; Cell Marque), desmin (D33, dilution 1:200; Dako), vimentin (V9, dilution 1:10 000; Biogenex), cytokeratin cocktails CK (AE1/AE3, dilution 1.200; Dako—CAM 5.2, dilution 1.50; Becton/Dickinson—Polyclonal, dilution 1:1000; Dako), MIB1(SP6, dilution 1:200; Labvision), L1 (IgG, Clone UJ127, dilution 1:50; NeoMarkers), p16 (dilution 1:500; MTM Laboratories AG, Heidelberg, Germany) and PKC theta (pT538 clone 19, dilution 1:20; BD Transduction Labs). The immunostaining was performed with an avidin–biotin detection system. Diaminobenzidine hydrochloride solution with hydrogen peroxide (Ventana Gen II, Dab basic) was the chromogen. Clinical positive control cases for all markers were included in every staining batch. Tissue cores were scored on the basis of the percentage of positive tumor cells staining above background, as completely negative, weakly stained (<10%), moderate positive (10–50%) or strongly positive (>50%).
Scores were entered into a Microsoft Excel spreadsheet. Scoring results were dichotomized into either negative (score of 0 or 1) or positive (score of 2 or 3) categories, and uninterpretable results were eliminated from further considerations (ie, they were not used for calculations for sensitivity and specificity). Discrepant score results for duplicate cores, when present, were consolidated as the higher interpretable score.27 To aid the analysis of the numerous tissue cores stained by immunohistochemistry, digital images were collected using a BLISS instrument (Bacus Laboratories, Lombard, IL, USA; http://bacuslabs.com). Scoring results were combined using Deconvoluter and represented in Treeview.28
Statistical Analysis
SPSS for Windows (version 14.0) was used for statistical analysis. Survival curves were plotted using Kaplan–Meier methods, with significance assessed using log-rank tests and based on overall survival over 20 years. Multivariate analyses were analyzed using Cox regression model. For correlations, the Pearson χ2-test was used.
Results
A total of 401 of the 651 samples represented in the tissue microarrays were positive for CD117 and regarded as definite GISTs. A total of 71% of these CD117-positive tumors stained positive for CD34, 19% for smooth muscle actin, 4% for desmin, <1% for S100 and 97% for vimentin. In addition, 37 cases not staining positive for CD117 or with cores missing from the CD117 tissue microarray slide (four cases) were positive for PKC theta. Four of these cases stained positive for S100 and were regarded to be true schwannomas. The remaining 33 cases were grouped together with the CD117-positive cases as immunohistochemically confirmed GISTs. Results of immunohistochemical staining are shown in Table 1. Clinicopathological results concerning GIST patients are presented in Table 2. No size was stated in 128 cases, but for the remaining cases, 20% of the tumors were classified into the very-low-risk and the low-risk groups, 34% in the intermediate-risk group and 47% in the high-risk group according to Fletcher et al.5 The standard markers (CD34, smooth-muscle actin, desmin or S100) did not correlate significantly with outcome.
A total of 182 of the 434 defined GISTs had 0 or 1 mitosis/50 high-power fields, 91 had 2–5, 45 patients had 6–10 and 116 had more than 10 mitoses/50 high-power fields. There was no significant difference in overall survival between the two groups with fewest mitoses, but for the group with 6–10 mitoses/50 high-power fields and again for the tumors with more than 10 mitoses/50 high-power fields, there was a significant less favorable overall survival (P=0.009 and 0.002). Median survival time for the patients with five mitoses or less was 6.5 years, for the group with 6–10 mitoses 3.2 years and for the patients with more than 10 mitoses 1.6 years. There was no significant correlation between the number of mitoses and location in the stomach or small bowel. Tumors with low mitotic index (<5/50 high-power fields) had a favorable overall survival (P<0.001) in both the locations.
Size of tumor has a close to linear curve, with a decrease in overall survival with increasing size. No significant difference was found however between tumors up to 2 cm and from 2 to 5 cm. Between groups of tumors from 5 to 10 cm and larger than 10 cm, there was likewise no difference. Patients with tumors 5 cm or less (108 cases) had a significantly better overall survival than those with tumors larger than 5 cm (198 cases), with a median survival time of 7.3 vs 3.2 years, respectively (P<0.001). There was a correlation between the number of mitoses and size, as tumors with more than five mitoses were found almost four times more frequently among tumors larger than 5 cm (P<0.001). There was a significant correlation between size and location, with 60% of the gastric tumors being 5 cm or larger compared with 75% of the small bowel tumors (P=0.011). The mean size of the gastric tumors was 8.7 cm and that of the small bowel tumors was 9.4 cm. Smaller size tumors gave a significantly better overall survival in gastric tumors (P<0.001), but not in the small bowel tumors (P=0.974).
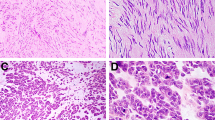
For patients with tumors without evident cellular atypia, the median survival time was 6.5 years compared with the group with focal or diffuse atypia (examples in Figure 1), where median survival was 2.4 and 1.8 years (P<0.001). Patients with gastric GISTs survived for 5.3 years compared with patients with small bowel tumors who survived for a median of 3.2 years (P=0.011). Females had an overall significantly better survival than men (median survival time 4.5 years vs 2.9 years, P=0.009). Other assessed parameters (ulceration, mucosal invasion, hemorrhage or necrosis) had no impact on overall survival.
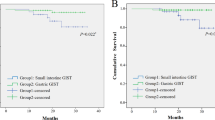
In our series, 180 (42%) of the defined GIST cases stained for p16, and these patients had a significantly worse overall survival (P=0.013) than patients not expressing p16 (Figure 2). Location of p16-expressing tumors is shown in Table 3. When evaluating only gastric tumors and tumors from the small bowel, the expression of p16 was still a negative prognostic factor compared with the group not expressing p16 (P=0.002). The distribution between gastric and small bowel tumors in the p16-expressing group and the group not expressing p16 shows significant differences (P=0.004). The number of gastric vs small bowel tumors was equal in the p16-expressing group, and with twice as many gastric tumors as small bowel tumors in the group not expressing p16. When investigating p16 in the group with gastric tumors, the patients not expressing p16 had an overall better survival (median survival time 6.5 years) compared with those who express p16 (median survival time 3.8 years, P=0.033). For patients with small bowel tumors, the median survival time for patients not expressing p16 was 5.2 years and for those expressing p16 was 3.2 years (P=0.065).
More than five mitoses/50 high-power fields correlated with tumors expressing p16, as 43% of these tumors had a high mitotic index. A total of 33% of the tumors not expressing p16 had more than five mitoses/50 high-power fields (P=0.050). Tumors expressing p16 were larger than 5 cm in 72% of cases, compared with those not expressing p16, where 59% of tumors were larger than 5 cm (P=0.021).
In a multivariate model evaluating all cases with complete data (N=306), the expression of p16 was a significant factor when mitotic index and size were accounted for (P=0.034).
A total of 255 (59%) of the GISTs expressed L1, and there was no difference in overall survival between the groups expressing or not expressing L1 (P=0.943) (Figure 3). This was also true for tumors located in the stomach or in the small bowel (P=0.074 and 0.184). L1 expression was more than three times as frequent among tumors located in the stomach compared with the small bowel tumors. For the tumors not expressing L1, only half of them were located in the stomach (P<0.001).
There was a significant correlation between tumors with more than five mitoses and the expression of L1 (P=0.043), with more than twice as many L1-expressing tumors having a high mitotic index than tumors not expressing L1. No correlation was found with tumor size. Location of L1-expressing tumors is shown in Table 3.
Discussion
This tissue microarray series was built using material from Norwegian pathology department archives. The series was based on tumors reported to the Cancer Registry of Norway and therefore tumors such as leiomyomas considered totally benign at the time of primary diagnosis were not registered. When comparing the numbers of patients in the different risk groups, our results showed less than half the number of cases in the very-low and low-risk groups, compared with a Swedish study described as being population based.9 The intermediate-risk group contained around 30% more patients in our study, but the high-risk group contained 25% more patients than in the Swedish study. Most other series have confined analyses to KIT-positive tumors, but we have also added PKC theta-positive cases in our study, based on recent reports by others,7, 8 which suggest that KIT-negative GISTs express PKC theta. Some of the KIT and PKC-double-negative cases can be categorized as leiomyomas, leiomyosarcomas, schwannomas or carcinomas based on other immunostains.
In GISTs, tumor size and mitotic activity are the most consistent clinicopathological features indicative of aggressive clinical behavior, features not readily assessable from typical bite or core biopsies. Here we have studied the immunohistochemical stainings for the proposed biomarkers p16 and L1 in GISTs to see if they can aid in the clinical prognostic assessment.
Decreased expression of p16 by immunohistochemical staining was previously reported to have a negative association with survival in GIST. The largest previous study on prognostic value of p16 in GISTs used patient material from five different pathology departments.13 Patients with tumors showing loss of p16 expression in their study had worse prognosis than those expressing p16. Our current larger study did not validate these findings, as patients expressing p16 were found to do worse than those not expressing this biomarker. Loss of p16 expression is biologically expected to contribute to malignancy. There are however many other oncogenic changes that can achieve the same net effect of increased proliferation, including loss of RB or TP53, and aberrant activation of cyclin D by gene amplification or by mutations activating the receptor tyrosine kinase-ras-raf-ERK-signaling cascade. Indeed, such alternative changes in cell-cycle control could lead to a compensatory upregulation of p16, and in support of this hypothesis we found p16 expression was actually more common among GISTs with higher mitotic counts. In cervical neoplasms, p16 is found to be increased in carcinoma in situ relative to benign lesions, and advanced carcinomas retain positivity for p16.29 In breast cancer, expression of p16 (rather than lack of expression) was found to correlate with poorer outcome.16
L1 is a protein with several interesting biological roles. In L1-mutant mice, corticospinal axons fail to project to the spinal cord.20 Anti-L1-antibodies cause alterations of branching morphogenesis in renal epithelium, leading to marked reduction of kidney size.21 L1 is expressed at the invasive front of colon cancers.30 Overexpression of L1 is associated with metastasis in cutaneous malignant melanomas, whereas inactivation of L1 significantly reduces the invasion and migration of melanoma cells.31 In the work of Keifi et al,25 L1 was evaluated both as a diagnostic and as a prognostic marker for GIST. Their study included 72 cases, with little information provided about the patient population from which the series was derived. A total of 73% of their GIST cases were L1 positive, and survival analysis was performed on 55 cases. Their analysis demonstrated only a trend to poorer survival in L1-positive vs L1-negative tumors (P=0.13), but has nevertheless been cited as supporting L1 as a factor for less favorable prognosis.22 We found no difference in survival between GISTs expressing L1 vs those not expressing L1.
Initially reported studies of a biomarker may show great promise, but subsequent studies on the same or related markers can yield inconsistent conclusions or sometimes even be in direct contradiction to the original reports. This has been recently discussed in a paper by McShane et al, where recommendations for tumor marker prognostic studies have been made.32 This article was simultaneously published in five journals, and presents guidelines for complete reporting so that relevant information is available for the readers to be able to judge the usefulness of the data. In retrospective studies, patient population is often biased toward patients with available tumor specimens. Specimen availability may be related to tumor size and patient outcome. In prospective studies the validation of prognostic factors can be confounded by patient selection. These problems are suggested as being surmountable by improvements in study design and reporting.33
We use a large series comprised of cases completely independent of the previous reports to be validated. Our results validate the established clinical prognostic markers, but do not validate the prognostic values of p16 and L1 immunohistochemical biomarkers presented in other series.
References
Miettinen M, Sarlomo-Rikala M, Lasota J . Gastrointestinal stromal tumours. Ann Chir Gynaecol 1998;87:278–281.
Steigen SE, Eide TJ . Trends in incidence and survival of mesenchymal neoplasm of the digestive tract within a defined population of northern Norway. APMIS 2006;11:192–200.
Miettinen M, Sobin LH, Lasota J . Gastrointestinal stromal tumors of the stomach: a clinicopathologic, immunohistochemical, and molecular genetic study of 1765 cases with long-term follow-up. Am J Surg Pathol 2005;29:52–68.
Miettinen M, Makhlouf H, Sobin LH, et al. Gastrointestinal stromal tumors of the jejunum and ileum: a clinicopathologic, immunohistochemical, and molecular genetic study of 906 cases before imatinib with long-term follow-up. Am J Surg Pathol 2006;30:477–489.
Miettinen M, Sobin LH, Sarlomo-Rikala M . Immunohistochemical spectrum of GISTs at different sites and their differential diagnosis with a reference to CD117 (KIT). Mod Pathol 2000;13:1134–1142.
Miettinen M, Lasota J . Gastrointestinal stromal tumors—definition, clinical, histological, immunohistochemical, and molecular genetic features and differential diagnosis. Virchows Arch 2001;438:1–12.
Blay P, Astudillo A, Buesa JM, et al. Protein kinase C theta is highly expressed in gastrointestinal stromal tumors but not in other mesenchymal neoplasias. Clin Cancer Res 2004;10:4089–4095.
Motegi A, Sakurai S, Nakayama H, et al. PKC theta, a novel immunohistochemical marker for gastrointestinal stromal tumors (GIST), especially useful for identifying KIT-negative tumors. Pathol Int 2005;55:106–112.
Nilsson B, Bumming P, Meis-Kindblom JM, et al. Gastrointestinal stromal tumors: the incidence, prevalence, clinical course, and prognostication in the preimatinib mesylate era—a population-based study in western Sweden. Cancer 2005;103:821–829.
Fletcher CD, Berman JJ, Corless C, et al. Diagnosis of gastrointestinal stromal tumors: a consensus approach. Hum Pathol 2002;33:459–465.
Kononen J, Bubendorf L, Kallioniemi A, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med 1998;4:844–847.
Hsu FD, Nielsen TO, Alkushi A, et al. Tissue microarrays are an effective quality assurance tool for diagnostic immunohistochemistry. Mod Pathol 2002;15:1374–1380.
Schneider-Stock R, Boltze C, Lasota J, et al. Loss of p16 protein defines high-risk patients with gastrointestinal stromal tumors: a tissue microarray study. Clin Cancer Res 2005;11:638–645.
Haller F, Gunawan B, von Heydebreck A, et al. Prognostic role of E2F1 and members of the CDKN2A network in gastrointestinal stromal tumors. Clin Cancer Res 2005;11:6589–6597.
Sabah M, Cummins R, Leader M, et al. Loss of heterozygosity of chromosome 9p and loss of p16INK4A expression are associated with malignant gastrointestinal stromal tumors. Mod Pathol 2004;17:1364–1371.
Munot K, Bell SM, Lane S, et al. Pattern of expression of genes linked to epigenetic silencing in human breast cancer. Hum Pathol 2006;37:989–999.
O'Neill CJ, McCluggage WG . p16 expression in the female genital tract and its value in diagnosis. Adv Anat Pathol 2006;13:8–15.
Moos M, Tacke R, Scherer H, et al. Neural adhesion molecule L1 as a member of the immunoglobulin superfamily with binding domains similar to fibronectin. Nature 1988;334:701–703.
Rathjen FG, Rutishauser U . Comparison of two cell surface molecules involved in neural cell adhesion. EMBO J 1984;3:461–465.
Cohen NR, Taylor JS, Scott LB, et al. Errors in corticospinal axon guidance in mice lacking the neural cell adhesion molecule L1. Curr Biol 1998;8:26–33.
Debiec H, Christensen EI, Ronco PM . The cell adhesion molecule L1 is developmentally regulated in the renal epithelium and is involved in kidney branching morphogenesis. J Cell Biol 1998;143:2067–2079.
Huszar M, Moldenhauer G, Gschwend V, et al. Expression profile analysis in multiple human tumors identifies L1 (CD171) as a molecular marker for differential diagnosis and targeted therapy. Hum Pathol 2006;37:1000–1008.
Thies A, Schachner M, Moll I, et al. Overexpression of the cell adhesion molecule L1 is associated with metastasis in cutaneous malignant melanoma. Eur J Cancer 2002;38:1708–1716.
Fogel M, Gutwein P, Mechtersheimer S, et al. L1 expression as a predictor of progression and survival in patients with uterine and ovarian carcinomas. Lancet 2003;362:869–875.
Kaifi JT, Strelow A, Schurr PG, et al. L1 (CD171) is highly expressed in gastrointestinal stromal tumors. Mod Pathol 2006;19:399–406.
Parker RL, Huntsman DG, Lesack DW, et al. Assessment of interlaboratory variation in the immunohistochemical determination of estrogen receptor status using a breast cancer tissue microarray. Am J Clin Pathol 2002;117:723–728.
Liu CL, Montgomery KD, Natkunam Y, et al. TMA-Combiner, a simple software tool to permit analysis of replicate cores on tissue microarrays. Mod Pathol 2005;18:1641–1648.
Liu CL, Prapong W, Natkunam Y, et al. Software tools for high-throughput analysis and archiving of immunohistochemistry staining data obtained with tissue microarrays. Am J Pathol 2002;161:1557–1565.
Cameron RI, Maxwell P, Jenkins D, et al. Immunohistochemical staining with MIB1, bcl2 and p16 assists in the distinction of cervical glandular intraepithelial neoplasia from tubo-endometrial metaplasia, endometriosis and microglandular hyperplasia. Histopathology 2002;41:313–321.
Gavert N, Conacci-Sorrell M, Gast D, et al. L1, a novel target of beta-catenin signaling, transforms cells and is expressed at the invasive front of colon cancers. J Cell Biol 2005;168:633–642.
Meier F, Busch S, Gast D, et al. The adhesion molecule L1 (CD171) promotes melanoma progression. Int J Cancer 2006;119:549–555.
McShane LM, Altman DG, Sauerbrei W, et al. REporting recommendations for tumour MARKer prognostic studies (REMARK). Eur J Cancer 2005;41:1690–1696.
Hayes DF . Prognostic and predictive factors revisited. Breast 2005;14:493–499.
Acknowledgements
We thank Dr David Owen for helpful advice and for providing additional expert review on 16 cases. TON is a Scholar of the Michael Smith Foundation for Health Research. The tissue microarray facility at the Genetic Pathology Evaluation Centre is supported in part by an unrestricted educational grant from sanofi-aventis. We also thank Kristin Smistad Myrmel for help with immunohistochemical scoring of the tissue microarray slides.
Author information
Authors and Affiliations
Corresponding author
Additional information
Conflict of interest statement
None declared.
Rights and permissions
About this article
Cite this article
Steigen, S., Bjerkehagen, B., Haugland, H. et al. Diagnostic and prognostic markers for gastrointestinal stromal tumors in Norway. Mod Pathol 21, 46–53 (2008). https://doi.org/10.1038/modpathol.3800976
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.3800976
Keywords
This article is cited by
-
Impact of age and gender on tumor related prognosis in gastrointestinal stromal tumors (GIST)
BMC Cancer (2015)
-
Spontaneous perforation of jejunal gastrintestinal stromal tumour (gist). Case report and review of literature
World Journal of Emergency Surgery (2012)
-
Tyrosine-kinase mutations in c-KIT and PDGFR-alpha genes of imatinib naïve adult patients with gastrointestinal stromal tumours (GISTs) of the stomach and small intestine: relation to tumour-biological risk-profile and long-term outcome
Clinical and Translational Oncology (2012)
-
p16Ink4a overexpression in cancer: a tumor suppressor gene associated with senescence and high-grade tumors
Oncogene (2011)
-
Gastrointestinal stromal tumors
Virchows Archiv (2010)