Abstract
Pleuropulmonary blastomas are rare malignant intrathoracic tumors of early childhood. They appear as a pulmonary- and/or pleural-based mass and their pathogenesis and relationship to other pediatric solid tumors is not well understood. In this study, paraffin-embedded material of five cases of pleuropulmonary blastoma was analyzed for genetic alterations by comparative genomic hybridization and five genetic loci by fluorescence in situ hybridization. Comparative genomic hybridization identified aberrations in all pleuropulmonary blastomas, including four amplifications in three tumors at chromosomes 5q33–34, 11q22.2–ter, 15q25–ter, and 19q11–13.2. The most frequent DNA gains involved 8q11–22.2 (four cases) and 20q (two cases), whereas the most common losses included 9p21–24 (two cases) and 11p14 (three cases). Chromosome 8 gains were confirmed by fluorescent in situ hybridization, resulting in the detection of up to five copies of chromosome 8 centromeres per nucleus. In the two surviving patients, chromosome 8 gains were the only genetic abnormality, suggesting that this might be an early event in pleuropulmonary blastoma carcinogenesis. The identification of new genetic alterations as well as the confirmation of previously reported ones (especially 8q gains) in pleuropulmonary blastoma should help to improve our understanding of both the molecular mechanisms underlying the tumorigenesis of pleuropulmonary blastoma and the relationship of pleuropulmonary blastoma with other pediatric tumors.
Similar content being viewed by others
Main
Pleuropulmonary blastomas are uncommon solid and/or cystic intrathoracic childhood tumors, which are considered part of the group of dysontogenetic tumors, such as Wilms’ tumors, hepatoblastomas, and neuroblastomas. Since their original description in 1988, they have been recognized as a distinct entity from adult pulmonary blastoma. The latter tumor consists of malignant epithelial and mesenchymal components, whereas in pleuropulmonary blastoma only the mesenchymal component is malignant, the epithelial component being regarded as entrapped elements.1 The mesenchyme may be undifferentiated or may show rhabdomyomatous, chondroid, or fibrosarcomatous differentiation. Generally, three gross types of pleuropulmonary blastoma have been distinguished: type 1, consisting entirely of multiple cysts; type 2, consisting of solid and cystic areas; and type 3, entirely composed of solid tumor tissue.2 Because of the cystic nature of pleuropulmonary blastoma types 1 and 2, a relationship has been suggested with another cystic lung lesion, congenital pulmonary airway malformation, although there appear to be clear clinical, histological, and genetic differences.3, 4 Patients are usually diagnosed between birth and 5 years of age and present relatively late with recurrent respiratory symptoms and chest pain.5 These tumors carry a poor prognosis, with a 42–49% 5-year survival rate, despite aggressive surgical and chemotherapeutic treatment.5, 6, 7 In addition, in 25% of pleuropulmonary blastoma patients or their family members other dysplasias or neoplasias occur. The diseases found to be associated with pleuropulmonary blastoma included cystic lung disease, cystic nephromas, Wilms’ tumors, sarcomas, medulloblastomas, thyroid neoplasias, malignant germ cell tumors, Hodgkin's disease, leukemia, and Langerhans cell histiocytosis.8, 9, 10
Owing to the relative rarity of these tumors, little is known about their genetic basis. Cytogenetic studies have repeatedly reported trisomy of chromosome 8 or, more generally, gains of chromosome 8, in addition to complex genetic abnormalities.10, 11, 12, 13, 14, 15, 16, 17, 18, 19 Furthermore, trisomy of chromosome 2 has been reported by fluorescence in situ hybridization (FISH).20 However, altogether these studies have investigated no more than 17 cases of pleuropulmonary blastoma. Apart from one case report,21 no studies have been performed using comparative genomic hybridization (CGH), a technology that allows genome-wide analysis of DNA copy number changes in tumor samples and which has a higher resolution than conventional karyotyping.22, 23, 24, 25 Here, we describe five patients with pleuropulmonary blastomas which have been analyzed by CGH and FISH. We show that pleuropulmonary blastomas contain multiple chromosomal abnormalities, including the previously described gain of chromosome 8, now further delineated to the long arm of chromosome 8, as well as heretofore undescribed losses and gains and, interestingly, several amplified chromosomal regions.
Materials and methods
Patients
All clinical data of our series of five patients have been summarized in Table 1. The patients were collected in a 10-year period from two pediatric oncology centers in The Netherlands (Rotterdam, Nijmegen) and one in Belgium (Antwerp). All patients were treated surgically, resulting in complete resection in four and incomplete resection of the tumor in the 28-month-old boy (patient 2). Four patients received additional chemotherapy according to the malignant mesenchymal tumor treatment protocol of the International Society for Pediatric Oncology (SIOP). In the remaining patient (patient 2), three preoperative and four postoperative rounds of chemotherapy (ifosfamide, vincristin, and actinomycin (IVA)) were given, followed by five rounds of cisplatinum and adriamycin after detection of residual disease. The father of one of the patients had had a multicystic renal lesion during childhood (although no slides are available for review, this most probably represents a cystic nephroma, which is known to be associated with pleuropulmonary blastoma), whereas the family history of the other four patients did not reveal any other relevant diseases.
Tissues
Formalin-fixed, paraffin-embedded tissue blocks were retrieved from the files of the Department of Pathology of the respective hospitals (see above), and hematoxylin and eosin (HE)-stained slides were reviewed by two pathologists (RRdK and AJMvU). All initial diagnoses were confirmed and the tumors were classified into pleuropulmonary blastoma types 1, 2, and 3, according to the gross specimen description and microscopic analysis.1 Routine immunohistochemical analysis was performed on 4-μm thick tissue sections from a representative block of each of the tumors, using antibodies to myoglobin, myogenin, vimentin, pankeratin, actin, CD34, smooth muscle actin, desmin, S100, and MyoD1. Incubation of slides was performed using a standard avidin–biotin–complex method. All primary antibodies were incubated for 30 min at room temperature, except MyoD1, which was incubated for 90 min. The technical details of the antibodies and the incubations are summarized in Table 2.
DNA Extraction
Genomic DNA from paraffin-embedded tumor blocks was isolated as described previously.25 Briefly, 4-μm thick HE-stained sections were used to select areas with more than 70% tumor cells. Selected parts that were easily recognized were scraped with a scalpel from glass slides with unstained 10-μm thick sections and collected in 1.5 ml Eppendorf tubes containing 100% ethanol. This material was spun down (5 min 14 000 r.p.m.) and dissolved in 1 ml xylol. These steps were repeated two times, after which the tissue samples were treated subsequently with 100 and 70% ethanol, with centrifugation steps in between (5 min 14 000 r.p.m.), and were air-dried prior to proteinase K digestion. Tissue collected in 1.5 ml Eppendorf tubes was digested overnight with 1 mg/ml Proteinase K (Sigma) in digestion buffer (50 mM Tris pH 8.0, 1 mM EDTA, 0.5% SDS) at 55°C. The DNA was purified either by using the QiaAmp DNA mini kit (Qiagen, Hilden, Germany) according to the manufacturer's recommendations or standard phenol–chloroform (PCI; Fluka) extraction, followed by ethanol precipitation and quantification by spectrophotometry.
CGH and Digital Image Analysis
CGH was performed essentially as described earlier.24, 25 Briefly, 2 μg tumor DNA was labeled with Spectrum Green-dUTPs (Vysis, Downers Grove, IL, USA) by nick translation (BioNick kit, Life Technologies, Basel, Switzerland). Spectrum Red-labeled normal reference DNA (Vysis) was used for co-hybridization. The hybridization mixture consisted of 800 ng Spectrum Green-labeled tumor DNA, 800 ng of Spectrum-Red labeled normal reference DNA and 10–15 μg of human Cot-1 DNA (Life Technologies) dissolved in 12 μl of hybridization buffer (50% formamide, 2 × SSC, pH 7.0, 10% dextrane sulfate). Hybridization was carried out for 3 days at 37°C to denatured (2 × SSC and 70% formamide, 2 × SSC, pH 7, each for 5 min at 75°C) normal male human metaphase spreads (Vysis). Slides were washed at 45°C two times for 5 min in 50% formamide/2 × SSC followed by dehydration in an ethanol series. The chromosomes were counterstained with 0.2 μg/ml DAPI in Vectashield for identification.
Digital images were recorded from 6 to 10 metaphases using the Metasystems Image Pro System (black and white CCD camera; Sandhausen, Germany) mounted on top of a Leica DM-RE fluorescence microscope equipped with appropriate filter sets for DAPI, Spectrum Green and Spectrum Red fluorescence. The images were analyzed using Isis 4.4.25 CGH software (Metasystems) to calculate average green to red ratio profiles for each chromosome. At least 10 observations per autosome and 5 observations per sex chromosome were included in each analysis. To confirm the reproducibility of the detected chromosomal abnormalities, CGH was also performed with reversed fluorochrome labels for tumor and reference DNA. Gains and losses of DNA sequences were defined as chromosomal regions where both the mean green-to-red fluorescence ratio and its s.d. were above 1.20 and below 0.80, respectively. Over-representations were considered amplifications when the fluorescence ratio values in a region of a chromosomal arm exceeded 1.5. In negative control hybridizations, the mean green-to-red ratio occasionally exceeded the fixed 1.2 cutoff level at the following chromosomal regions: 1p32–pter, 16p, 19, and 22. Gains of these known G-C-rich regions were therefore excluded from all analyses.
Confirmation of CGH Data by FISH
To validate CGH data independently, paraffin-embedded tissue sections (4 μm) of the pleuropulmonary blastomas were subjected to FISH and evaluated as described previously.25 For this purpose, repetitive, fluorochrome-labeled DNA probes specific for human centromeres 1, 2, 8, X, and Y (in the male patient) were used. The pleuropulmonary blastoma showing a 19q11–13.2 amplification by CGH was moreover hybridized with an AKT gene-specific BAC probe.
Results
Histology and Immunohistochemistry
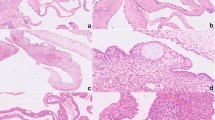
All tumors showed a histological picture compatible with pleuropulmonary blastoma. In types 1 and 2 there were cysts lined by non-tumoral flattened to cuboidal epithelium, underneath which there was a cambium-like layer of atypical mesenchymal cells with a variable mitotic frequency. Cellularity varied from high to low, and sometimes there were areas with epithelioid mesenchymal cells, mostly in more solid parts, which predominated in two tumors (patients 3 and 4). In these two patients, there was also chondroid differentiation in the solid areas. Immunohistochemistry yielded similar staining results in all tumors, with myogenin, vimentin, and desmin being diffusely and strongly positive, except for one case (patient 4) where there was only focal desmin and myogenin staining, which might be related to the fact that this was a type 3 (solid) pleuropulmonary blastoma. Furthermore, there was focal staining of varying intensity for myoglobin, MyoD1, and actin, again with the type 3 pleuropulmonary blastoma showing the least staining for MyoD1 and actin. Finally, pankeratin, CD34, smooth muscle actin, and S100 could not be detected in tumor cells. Representative pictures of the immunostainings are shown in Figure 1.
Overview of CGH Findings
Table 3 summarizes all DNA copy number changes identified in the five pleuropulmonary blastomas. Genetic alterations were observed in all tumors, and the average number of chromosome arm aberrations per tumor was 6.6 (range: 1–12). Both losses and gains were identified, as well as four amplifications in three tumors. The two surviving patients showed only gain of chromosome 8 material and had no amplifications. The most common chromosome arm copy number change was gain of the long arm of chromosome 8, which occurred in four tumors (see Figure 2a). Furthermore, two pleuropulmonary blastomas showed gain of the long arm of chromosome 20, whereas loss of the short arm of chromosomes 9 and 11 were identified in two and three tumors, respectively. Interestingly, there were 4 amplifications in three tumors, including chromosomes 5q33–34, 11q22.2–ter, 15q25–ter, and 19q11–13.2. This latter amplification is shown in Figure 3.
Representative comparative genomic hybridization (CGH) picture with corresponding confirmatory fluorescence in situ hybridization (FISH) analysis of chromosome 8q gain, showing the CGH result in the right upper panel (a), and the result of the FISH analysis, with a probe for the centromere of chromosome 8, in the right lower panel (b). There is a clear increase in the number of FISH spots, resulting in up to five copies per nucleus. For comparison, the CGH result of chromosome 1 and the corresponding FISH experiment with a probe to the centromere of chromosome 1 are shown in the left upper and lower panel, respectively. In the FISH experiment, only two copies of chromosome 1 are present.
Comparative genomic hybridization (CGH) result for chromosome 19q of patient 3, showing amplification of 19q11–13.2 in the right upper panel (a). The corresponding fluorescence in situ hybridization (FISH) analysis, using a probe for AKT, is depicted in the right lower panel (b), in which many nuclear spots of high intensity can be appreciated, as a result of the amplification. For comparison, the CGH result and corresponding FISH analysis for one of the other patients (patient 1) have been depicted in the corresponding left upper and lower panel, respectively, showing two weaker signals per nucleus.
Comparison of CGH and FISH Results
FISH results with four centromere and one AKT-specific probes are listed in Table 3. We could confirm the CGH results of chromosome 8q gain in all tumors that were analyzed by FISH, which detected up to five copies of chromosome 8 centromeres per nucleus, as is shown in Figure 2b. Also, the amplification of 19q11–13.2 was confirmed by FISH analysis of the AKT-specific probe, as is shown in Figure 3b.
Discussion
In this study, we have performed CGH analysis on a series of five pleuropulmonary blastomas and shown that there are several areas of gain (8qcen-22.2 and 20q) and loss (9p21–24 and 11p14) of chromosomal material, and several amplifications, including 5q33–34, 11q22.2–ter, 15q25–ter, and 19q11–13.2. The gain of 8q and the amplification of 19q11–13.2 were confirmed by FISH analysis. In contrast to earlier papers, no abnormalities in chromosome 2 were found. The areas with gains and losses may be indications for the presence of oncogenes or tumor suppressor genes, respectively, and may direct further investigations to increase our understanding of the molecular pathogenesis of these rare tumors.
So far, molecular genetic abnormalities in pleuropulmonary blastomas have been investigated in single cases or in small series of tumors, due to the rare occurrence. The most consistent chromosomal anomaly is extra copies of chromosome 8, which has been found in many studies, approximating 75–80% of all cases studied.10, 11, 12, 13, 14, 15, 16, 17, 18, 19 A single case study has thus far been performed with CGH, also showing gain of almost the entire chromosome 8.21 In our study, we confirm these findings by showing gain of at least the long arm of chromosome 8 in four of the five cases studied. Interestingly, the two surviving patients in our series (patients 1 and 5), both had a gain of chromosome 8 material as the only genetic abnormality. Although our series is too small to draw definitive conclusions, it appears that type 1 or 2 pleuropulmonary blastomas histology presents with few genetic changes, whereas type 3 histology presents with more advanced cancer, characterized by an accumulation of genetic changes and a worse prognosis.
We were able to pinpoint the smallest region of overlap in our four cases to 8q11.1–22.2. While strongly indicating that a gene initiating pleuropulmonary blastoma tumorigenesis may be located here, this region is still large and contains many candidate genes. Interestingly, the MYC and DDEF1 oncogenes can be excluded as candidates, as these genes are located on 8q24. Conversely, the MOS oncogene, located on 8q11, might play a role in pleuropulmonary blastoma tumorigenesis, as has also been postulated in astrocytic tumors of the brain and in non-small cell lung cancer.26, 27 The only additional chromosomal region with chromosomal gain in more than one patient was 20q. Its importance in pleuropulmonary blastomas, however, is unclear because this gain has been described before in only one patient.
In contrast to the gains, which involved large parts of chromosomes, the identified losses comprised relatively limited regions of smallest overlap, indicating loci harboring putative tumor suppressor genes. Within 9p21–24, which was lost in two of the pleuropulmonary blastomas investigated, the most relevant candidate tumor suppressor genes appear to be CDKN2A (p16INK4A), which inhibits the action of cyclin-dependent kinase 4 (CDK4), and CDKN2B (p14ARF), which inhibits the action of MDM2.28 The inactivation of these genes by loss, mutation, or hypermethylation are key events in a variety of tumors, including melanomas, gastrinomas, colorectal carcinoma, and non-small cell carcinomas of the lung.29, 30, 31, 32 On the short arm of chromosome 11 we were also able to pinpoint a relatively small region that was lost in three pleuropulmonary blastomas in our series, which consisted of 11p14 only. While this region does not appear to contain currently known tumor suppressor genes, it is interesting to note that two genes, WT1 and PAX6, that are highly relevant to pediatric tumorigenesis and development are located on 11p13, which is deleted in two patients. WT1 and PAX6 are involved in the pathogenesis of Wilms’ tumors and the WAGR (Wilms’ tumor, aniridia, genito-urinary abnormalities, mental retardation) syndrome.33, 34 In addition, 11p15.5, a chromosomal region known to harbor several imprinted genes, as well as genes also involved in pediatric tumorigenesis, is deleted in two patients. More specifically, several genes in this region have been implicated in the Beckwith–Wiedemann syndrome (BWS), such as CDKN1C and WT2. BWS is characterized by an increased frequency of childhood cancers, including Wilms’ tumors and rhabdomyosarcomas.35, 36 Thus, CDKN1C, encoding the CDK inhibitor 1C (p57KIP2) and WT2 might be considered relevant candidate genes in this region.37, 38 The fact that 11p13 containing WT1, and 11p15.5 containing WT2, are each lost in two of our patients, is even more relevant against the background of the association that has been reported between the occurrence of Wilms’ tumors and pleuropulmonary blastomas in the literature.8, 10
Losses at 9p and 11p in pleuropulmonary blastomas had been found in only one other study. It should be noted, however, that only one previous study had employed CGH on one patient. All other studies involved conventional or spectral karyotype analysis and reported predominantly gains or losses of entire chromosomes and a large variety of translocations and other chromosomal rearrangements in highly aneuploid or polyploid tumors. This may reflect more extensive chromosomal instability in more advanced tumors. However, it may well be that CGH is able to detect smaller regions of chromosomal gains and losses.
In addition to gains and losses, we found four amplifications that occurred in three tumors. One of these regions, 15q25–ter, was found to be gained in another recently reported pleuropulmonary blastoma case, but the other regions so far appear to be unique findings.18 With the exception of 5q33–34, which represents a relatively small region, all other amplified chromosomal regions are much larger and contain at least 200 genes, making selection of candidate genes difficult. Yet, a few genes could be regarded as such, including cyclin G1 on 5q, which appears to be involved in carcinogenesis in uterine leiomyomas and in an animal model of hepatic tumors.39, 40 The ETS1 proto-oncogene, encoding a nuclear transcription factor, is located on 11q23.3 and is involved in various types of human cancers, including ovarian cancer, endometrium cancer, and gastric cancer.41, 42, 43 Finally, 19q13.1–q13.2 is the locus for AKT2, an isoform of the phosphoinositide-dependent serine–threonine protein kinase Akt.44 While being responsible for a familial form of autosomal dominant type II diabetes mellitus when mutated, its overexpression has been found in various types of cancer and cancer cell lines, such as breast and ovarian cancer.45, 46, 47 Further cancer-related genes that might be considered in the same chromosomal region are PAK4, implicated in pancreatic cancer, and AXL, a receptor tyrosine kinase.
Taken together, we have analyzed five pleuropulmonary blastomas by CGH and shown that gain of the long arm of chromosome 8 is a frequent finding in pleuropulmonary blastoma tumorigenesis. Although still lacking support from this or other studies, the MOS oncogene appears one candidate gene to be studied in more detail. Further, we have found losses of 9p and 11p, both containing relevant candidate tumor suppressor genes to be further analyzed. While this study presents clues to unravel the molecular pathogenesis of pleuropulmonary blastomas, additional genome-wide studies with larger numbers of patients are required to confirm the results of the present study and to better understand this rare neoplasm.
References
Manivel JC, Priest JR, Watterson J, et al. Pleuropulmonary blastoma. The so-called pulmonary blastoma of childhood. Cancer 1988;62:1516–1526.
Dehner LP, Watterson J, Priest J . Pleuropulmonary blastoma. A unique intrathoracic pulmonary neoplasm of childhood. Perspect Pediatr Pathol 1995;18:214–226.
MacSweeney F, Papagiannopoulos K, Goldstraw P, et al. An assessment of the expanded classification of congenital cystic adenomatoid malformations and their relationship to malignant transformation. Am J Surg Pathol 2003;27:1139–1146.
Hill DA, Dehner LP, Ackerman LV . A cautionary note about congenital cystic adenomatoid malformation (CCAM) type 4. Am J Surg Pathol 2004;28:554–555.
Priest JR, McDermott MB, Bhatia S, et al. Pleuropulmonary blastoma: a clinicopathologic study of 50 cases. Cancer 1997;80:147–161.
Parsons SK, Fishman SJ, Hoorntje LE, et al. Aggressive multimodal treatment of pleuropulmonary blastoma. Ann Thorac Surg 2001;72:939–942.
Indolfi P, Bisogno G, Casale F, et al. Prognostic factors in pleuro-pulmonary blastoma. Pediatr Blood Cancer 2007;48:318–323.
Messinger YH, Dehner LP, Hill DA, et al. Pleuropulmonary blastoma and its association with other childhood neoplasms. J Clin Oncol 2004;22:8542.
Bouron-dal Soglio D, Harvey I, Yazbeck S, et al. An association of pleuropulmonary blastoma and cystic nephroma: possible genetic association. Pediatr Dev Pathol 2006;9:61–64.
Priest JR, Watterson J, Strong L, et al. Pleuropulmonary blastoma: a marker for familial disease. J Pediatr 1996;128:220–224.
Sciot R, Dal Cin P, Brock P, et al. Pleuropulmonary blastoma (pulmonary blastoma of childhood): genetic link with other embryonal malignancies? Histopathology 1994;24:559–563.
Novak R, Dasu S, Agamanolis D, et al. Trisomy 8 is a characteristic finding in pleuropulmonary blastoma. Pediatr Pathol Lab Med 1997;17:99–103.
Barnard M, Bayani J, Grant R, et al. Use of multicolor spectral karyotyping in genetic analysis of pleuropulmonary blastoma. Pediatr Dev Pathol 2000;3:479–486.
Sebire NJ, Rampling D, Malone M, et al. Gains of chromosome 8 in pleuropulmonary blastomas of childhood. Pediatr Dev Pathol 2002;5:221–222.
Vargas SO, Nose V, Fletcher JA, et al. Gains of chromosome 8 are confined to mesenchymal components in pleuropulmonary blastoma. Pediatr Dev Pathol 2001;4:434–445.
Hong B, Chen Z, Coffin CM, et al. Molecular cytogenetic analysis of a pleuropulmonary blastoma. Cancer Genet Cytogenet 2003;142:65–69.
Kelsey AM, McNally K, Birch J, et al. Case of extra pulmonary, pleuro-pulmonary blastoma in a child: pathological and cytogenetic findings. Med Pediatr Oncol 1997;29:61–64.
Maerken T van, Roy N van, Dhooge C, et al. Cytogenetic characterization of two pleuropulmonary blastoma cases: evidence for polysomy 8, 17p loss and distal 10q loss as recurrent aberrations. Pediatr Blood Cancer 2005;45:441.
Quilichini B, Andre N, Bouvier C, et al. Hidden chromosomal abnormalities in pleuropulmonary blastomas identified by multiplex FISH. BMC Cancer 2006;6:4.
Yang P, Hasegawa T, Hirose T, et al. Pleuropulmonary blastoma: fluorescence in situ hybridization analysis indicating trisomy 2. Am J Surg Pathol 1997;21:854–859.
Roque L, Rodrigues R, Martins C, et al. Comparative genomic hybridization analysis of a pleuropulmonary blastoma. Cancer Genet Cytogenet 2004;149:58–62.
Forozan F, Karhu R, Kononen J, et al. Genome screening by comparative genomic hybridization. Trends Genet 1997;13:405–409.
Knuutila S, Bjorkqvist AM, Autio K, et al. DNA copy number amplifications in human neoplasms: review of comparative genomic hybridization studies. Am J Pathol 1998;152:1107–1123.
Dannenberg H, Speel EJM, Zhao J, et al. Losses of chromosomes 1p and 3q are early genetic events in the development of sporadic pheochromocytomas. Am J Pathol 2000;157:353–359.
Speel EJM, Scheidweiler AF, Zhao J, et al. Genetic evidence for early divergence of small functioning and non-functioning endocrine pancreatic tumors: gain of 9q34 is an early event in insulinomas. Cancer Res 2001;61:5186–5192.
Zacharatos P, Kotsinas A, Tsantoulis P, et al. Relationship of the K-ras/c-mos expression patterns with angiogenesis in non-small cell lung carcinomas. Mol Med 2001;7:590–597.
Perunovic B, Athanasiou A, Quilty RD, et al. Expression of mos in astrocytic tumors and its potential role in neoplastic progression. Hum Pathol 2002;33:703–707.
Lilischkis R, Sarcevic B, Kennedy C, et al. Cancer-associated mis-sense and deletion mutations impair p16INK4 CDK inhibitory activity. Int J Cancer 1996;66:249–254.
Dracopoli NC, Fountain JW . CDKN2 mutations in melanoma. Cancer Surv 1996;26:115–132.
Ohhara M, Esumi M, Kurosu Y . Activation but not inactivation of the MTS1 gene is associated with primary colorectal carcinomas. Biochem Biophys Res Commun 1996;226:791–795.
Serrano J, Goebel SU, Peghini PL, et al. Alterations in the p16INK4a/CDKN2A tumor suppressor gene in gastrinomas. Clin Endocrinol Metab 2000;85:4146–4156.
Packenham JP, Taylor JA, White CM, et al. Homozygous deletions at chromosome 9p21 and mutation analysis of p16 and p15 in microdissected primary non-small cell lung cancers. Clin Cancer Res 1995;1:687–690.
Muto R, Yamamori S, Ohashi H, et al. Prediction by FISH analysis of the occurrence of Wilms tumor in aniridia patients. Am J Med Genet 2002;108:285–289.
Crolla JA, van Heyningen V . Frequent chromosome aberrations revealed by molecular cytogenetic studies in patients with aniridia. Am J Hum Genet 2002;71:1138–1149.
Steenman M, Westerveld A, Mannens M . Genetics of Beckwith-Wiedemann syndrome-associated tumors: common genetic pathways. Genes Chromosomes Cancer 2000;28:1–13.
Cohen Jr MM . Beckwith-Wiedemann Syndrome: Historical, Clinicopathological, and Etiopathogenetic Perspectives. Pediatr Dev Pathol 2005;8:287–304.
Hatada I, Ohashi H, Fukushima Y, et al. An imprinted gene p57KIP2 is mutated in Beckwith-Wiedemann syndrome. Nat Genet 1996;14:171–173.
Crider-Miller SJ, Reid LH, Higgins MJ, et al. Novel transcribed sequences within the BWS/WT2 region in 11p15.5: tissue-specific expression correlates with cancer type. Genomics 1997;46:355–363.
Baek WK, Kim D, Jung N, et al. Increased expression of cyclin G1 in leiomyoma compared with normal myometrium. Am J Obstet Gynecol 2003;188:634–639.
Jensen MR, Factor VM, Fantozzi A, et al. Reduced hepatic tumor incidence in cyclin G1-deficient mice. Hepatology 2003;37:862–870.
Yu Y, Zhang YC, Zhang WZ, et al. Ets1 as a marker of malignant potential in gastric carcinoma. World J Gastroenterol 2003;9:2154–2159.
Takai N, Miyazaki T, Nishida M, et al. c-Ets1 is a promising marker in epithelial ovarian cancer. Int J Mol Med 2002;9:287–292.
Takai N, Miyazaki T, Fujisawa K, et al. Expression of c-Ets1 is associated with malignant potential in endometrial carcinoma. Cancer 2000;89:2059–2067.
Cheng JQ, Godwin AK, Bellacosa A, et al. AKT2, a putative oncogene encoding a member of a subfamily of protein-serine/threonine kinases, is amplified in human ovarian carcinomas. Proc Natl Acad Sci USA 1992;89:9267–9271.
George S, Rochford JJ, Wolfrum C, et al. A family with severe insulin resistance and diabetes due to a mutation in AKT2. Science 2004;304:1325–1328.
Arboleda MJ, Lyons JF, Kabbinavar FF, et al. Overexpression of AKT2/protein kinase Bbeta leads to up-regulation of beta1 integrins, increased invasion, and metastasis of human breast and ovarian cancer cells. Cancer Res 2003;63:196–206.
Bellacosa A, de Feo D, Godwin AK, et al. Molecular alterations of the AKT2 oncogene in ovarian and breast carcinomas. Int J Cancer 1995;64:280–285.
Acknowledgements
We thank Dr Jack Priest for critically reading the manuscript and helpful comments. Further, we thank Ms Ineke Bouma for her help in preparing the Tables, Ms Esther Korpershoek for her aid in the analysis of the relevant chromosomal regions, Mr Frank van der Panne for photographic assistance, and Mr Pieter Derkx for the details of antibodies and the stainings.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
de Krijger, R., Claessen, S., van der Ham, F. et al. Gain of chromosome 8q is a frequent finding in pleuropulmonary blastoma. Mod Pathol 20, 1191–1199 (2007). https://doi.org/10.1038/modpathol.3800953
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.3800953
Keywords
This article is cited by
-
Metadherin (MTDH) overexpression significantly correlates with advanced tumor grade and stages among colorectal cancer patients
Molecular Biology Reports (2021)
-
FGF10 Signaling differences between type I pleuropulmonary blastoma and congenital cystic adenomatoid malformation
Orphanet Journal of Rare Diseases (2013)