Abstract
Dermatofibrosarcoma protuberans is a superficial low-grade sarcoma that rarely evolves into a high-grade fibrosarcoma. Dermatofibrosarcoma protuberans is genetically characterized by the unbalanced chromosomal t(17;22)(q21;q13), usually in the form of a supernumerary ring chromosome. The product of this chromosomal translocation is the chimeric gene COL1A1-PDGFB (collagen type I alpha I-platelet-derived growth factor beta), which is amplified at low levels in the ring chromosome. The aims of this study were to evaluate (1) whether genomic gains of this fusion gene occur during the clonal evolution of dermatofibrosarcoma protuberans into fibrosarcomatous dermatofibrosarcoma protuberans and (2) whether there is a difference between the number of genomic copies of COL1A1-PDGFB between classic dermatofibrosarcoma protuberans and dermatofibrosarcoma protuberans areas associated with fibrosarcomatous dermatofibrosarcoma protuberans. Eleven cases of fibrosarcomatous dermatofibrosarcoma protuberans with both dermatofibrosarcoma protuberans and fibrosarcomatous areas and 10 cases of classic dermatofibrosarcoma protuberans were studied. Genomic copies of COL1A1-PDGFB were evaluated by fluorescence in situ hybridization using a custom designed probe for the PDGFB locus on 4 μm thick paraffin-embedded tissue sections. Genomic gains of the COL1A1-PDGFB gene were observed in six (of 10) fibrosarcomatous dermatofibrosarcoma protuberans in the fibrosarcomatous areas when compared to the dermatofibrosarcoma protuberans areas of the same tumor (2–7 gene copies (median PDGFB copy gain, 2.8) versus 1–3 gene copies (median PDGFB copy gain, 1.7), respectively, P=0.004). Four fibrosarcomatous dermatofibrosarcoma protuberans did not show genomic gains of COL1A1-PDGFB fusion gene between the two areas. Essentially no difference in the copy number of COL1A1-PDGFB fusion gene was observed between dermatofibrosarcoma protuberans areas of classic dermatofibrosarcoma protuberans and dermatofibrosarcoma protuberans areas of fibrosarcomatous dermatofibrosarcoma protuberans (median PDGFB copy gain of 1.8 versus 1.7, respectively, P=0.36). Genomic gains of COL1A1-PDGFB fusion gene is possibly an oncogenic mechanism that is identified in the clonal evolution of a subset of dermatofibrosarcoma protuberans that evolves into fibrosarcomatous dermatofibrosarcoma protuberans. Since this finding was not observed in all cases of fibrosarcomatous dermatofibrosarcoma protuberans, other oncogenic mechanisms may be operating in this form of tumor progression. Copy number of COL1A1-PDGFB fusion gene in the classic dermatofibrosarcoma protuberans areas does not seem to be a major predisposing mechanism for fibrosarcomatous transformation.
Similar content being viewed by others
Main
Dermatofibrosarcoma protuberans is a locally aggressive dermal and subcutaneous tumor, which has very small risk of metastases.1 Fibrosarcomatous transformation rarely occurs in dermatofibrosarcoma protuberans and is associated with an increased risk for metastases.2, 3 Current treatment for dermatofibrosarcoma protuberans or fibrosarcomatous dermatofibrosarcoma protuberans consists of wide local excision with negative margins and possible adjuvant radiotherapy in selected cases.4, 5
Dermatofibrosarcoma protuberans is cytogenetically characterized by an unbalanced chromosomal translocation t(17;22)(q22;q13), usually presenting as a supernumerary ring chromosome containing genomic sequences of both chromosome 17 and 22.6, 7, 8 The result of this chromosomal translocation is the creation of a fusion gene with sequences from collagen type I alpha I (COL1A1) on chromosome 17q22 fused to the platelet-derived growth factor beta (PDGFB) on chromosome 22q13. The fusion product COL1A1-PDGFB is amplified at low levels on the ring chromosome (1–3 copies) and its protein product creates an autocrine loop with increased PDGFRB activity.9, 10, 11
Early case reports examining the cytogenetic alterations within fibrosarcomatous dermatofibrosarcoma protuberans indicate that the COL1A1-PDGFB fusion product is present in both fibrosarcomatous and dermatofibrosarcoma protuberans areas, indicating a common genetic origin for the two tumor components.12 However, a single previous study was not able to find a difference in copy numbers between fibrosarcomatous and dermatofibrosarcoma protuberans areas using comparative genomic hybridization.13 The objective of our study was to evaluate whether genomic gains of the COL1A1-PDGFB fusion gene occur during the clonal evolution of dermatofibrosarcoma protuberans into fibrosarcomatous dermatofibrosarcoma protuberans using fluorescence in situ hybridization. In addition, we aimed to determine if there was a difference between the number of COL1A1-PDGFB gene fusion copies between cases of classic dermatofibrosarcoma protuberans and dermatofibrosarcoma protuberans areas associated with fibrosarcomatous transformation.
Materials and methods
Patient Data
Eleven cases of fibrosarcomatous transformation of dermatofibrosarcoma protuberans, 10 of which with distinctly identifiable dermatofibrosarcoma protuberans and fibrosarcomatous areas, were examined from the extramural consultation files of the Department of Laboratory Medicine and Pathology at Mayo Clinic in Rochester, Minnesota, after Mayo Foundation Institutional Review Board approval. Ten cases of classic dermatofibrosarcoma protuberans were also examined from both intramural and extramural sources. In each case, 4 μm-thick sections of formalin-fixed, paraffin-embedded material were stained with hematoxylin and eosin and reviewed to confirm the diagnosis and correlate with fluorescence in situ hybridization results (see below).
Fluorescence In Situ Hybridization
BAC clones flanking the PDGFB locus on chromosome 22q13 were obtained from Children's Hospital Oakland Research Institute (Oakland, CA, USA). DNA isolation was performed according to Qiagen plasmid Maxi Kit specifications. DNA was directly labeled using nick translation kit from Abbot Laboratory using the Spectrum Orange and Spectrum Green fluorochromoes (Vysis Inc., Downers Grove, IL, USA).
Interphase molecular cytogenetic studies were performed on 4 μm paraffin-embedded thin tissue sections that were deparaffinized in xylene (2 × 15 min), dehydrated twice in 100% in ethylic alcohol for 5 min, and treated with 10 mmol/l citric acid for 10 min in a humid microwave. Tissue sections were then transferred to 37°C 2 × SSC for 5 min and protein digested with Digest All-3 (Zymed, San Francisco, CA, USA). After brief washing in 1 × PBS, the slides were sequentially dehydrated in alcohol (70, 85 and 100%) and air-dried at room temperature. Tissue sections were denatured at 80°C for 5 min and BAC probe hybridization was carried out overnight in a humidified chamber at 37°C. Tissue sections were then washed in 0.1% NP40/2 × SSC at 76°C for 4 min, then washed in 0.1% NP40/2 × SSC at room temperature for 1 min. Slides were then mounted in VACTASHIELD mounting medium with 1.5 μg/ml of 4′,6-diamidino-2-phenylindole. Tumor samples were scored by three investigators in approximately 300–1000 cells within both the dermatofibrosarcoma protuberans and fibrosarcomatous areas. Two different counting techniques were used: (1) green and orange signals were tallied in individual cells; and (2) signals were counted individually in a high power fields ( × 1000) and ratios of green to red signals were tallied. The COL1A1-PDGFB copy gain represents the average copy gain of the fusion gene per cell and was calculated by subtracting green signals from red signals and dividing by total number of cells counted.
Statistical Analysis
All group comparisons were performed using the Wilcoxon test. Paired analysis between dermatofibrosarcoma protuberans and fibrosarcomatous areas was performed with the Wilcoxon paired signed rank test. Level of significance was set at P≤0.05 (two tailed).
Results
Clinicopathologic Features
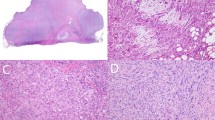
Clinical characteristics of the study group are listed in Table 1. On histologic examination (Figure 1), all tumors had a dermatofibrosarcoma protuberans component characterized by a uniform, low-grade spindle cell proliferation growing in a storiform pattern and in an infiltrative manner (Figure 1a). Mitotic counts in this area ranged from 0 to 2 mitoses/10 high-power fields. Case 9 also had a confirmed dermatofibrosarcoma protuberans component but the block was not available for further study. High-grade fibrosarcomatous areas were present in all 11 cases and consisted of markedly cellular areas with a herringbone pattern of growth and an increased mitotic rate (Figure 1b).
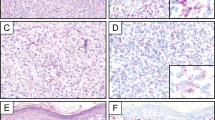
Case 7. (a) Excision of a superficial tumor from the back of an 85-year-old male shows a classic dermatofibrosarcoma protuberans; a cellular spindle cell tumor diffusely infiltrating the deep dermis and subcutaneous fat with formation of a storiform architectural pattern (H&E; original magnification × 100). (b) The fibrosarcomatous region from the same tumor shows a ‘herringbone’ fascicular architecture, increased cellularity, and increased mitotic activity (H&E; original magnification × 100). (c) Fluorescence in situ hybridization of the dermatofibrosarcoma protuberans component of the tumor using a custom designed breakapart probe to the PDGFB locus on chromosome 22q13 shows several tumor cells with 1 red–green signal indicating a normal chromosome 22 and 1–3 extra copies of red signal representing COL1A1-PDGFB fusion gene (arrows) (original magnification, × 1000). (d) Fluorescence in situ hybridization of the fibrosarcomatous component of the tumor using a custom designed breakapart probe to the PDGFB locus on chromosome 22q13 shows several tumor cells with 1 red–green signal indicating a normal chromosome 22 and 3–7 extra copies of red signal representing COL1A1-PDGFB fusion gene (arrow) (original magnification, × 1000).
Fluorescence In Situ Hybridization
Genomic gains of the COL1A1-PDGFB gene were observed in six (of 10) cases of fibrosarcomatous dermatofibrosarcoma protuberans in the fibrosarcomatous areas when compared to the classic dermatofibrosarcoma protuberans areas paired for individual cases (P=0.004) (Table 1 and Figure 2). The median COL1A1-PDGFB copy gain in dermatofibrosarcoma protuberans areas of fibrosarcomatous dermatofibrosarcoma protuberans was 1.7 (range 0.2–2.8) (Figure 1d) while the median COL1A1-PDGFB copy gain in fibrosarcomatous areas of fibrosarcomatous dermatofibrosarcoma protuberans was 2.8 (range 0–5.9) (Figure 1c). Four (of 10) cases of fibrosarcomatous dermatofibrosarcoma protuberans showed no difference in COL1A1-PDGFB copy gains between the fibrosarcomatous areas and dermatofibrosarcoma protuberans areas.
A very small difference in COL1A1-PDGFB copy number gains was observed in cases of classic dermatofibrosarcoma protuberans and the dermatofibrosarcoma protuberans areas of fibrosarcomatous dermatofibrosarcoma protuberans but the changes were not statistically significant (P=0.36) (Table 1 and Figure 2). The median COL1A1-PDGFB copy gain in classic dermatofibrosarcoma protuberans was 1.8 (range 0–2.4) while the median COL1A1-PDGFB copy gain in dermatofibrosarcoma protuberans areas of fibrosarcomatous dermatofibrosarcoma protuberans was 1.7 (range 0.2–2.8).
Metastatic Fibrosarcomatous Dermatofibrosarcoma Protuberans
A single case (case 11) of fibrosarcomatous dermatofibrosarcoma protuberans metastasized to distant bone and morphologically showed 100% fibrosarcomatous differentiation. The metastatic lesion possessed an average COL1A1-PDGFB copy number gain of 2.1, but this number was essentially the same as the 2.0 average copy number gain in the fibrosarcomatous area of the primary lesion (Table 1).
Discussion
Fibrosarcomatous change in dermatofibrosarcoma protuberans has been increasingly recognized as a form of tumor progression that carries an increased risk of metastasis over classic dermatofibrosarcoma protuberans, but2, 3, 14 the biologic mechanisms underlying this transformation are poorly understood. Prior studies have highlighted the role of increased proliferation rates and TP53 mutations in the pathogenesis of fibrosarcomatous change in dermatofibrosarcoma protuberans.2, 15, 16, 17 In addition, other studies on fibrosarcomatous dermatofibrosarcoma protuberans have shown that the COL1A1-PDGFB fusion gene is present in both dermatofibrosarcoma protuberans and fibrosarcomatous areas.6, 18 Wang et al12 detected the presence of the COL1A1-PDGFB fusion gene in five of six cases studied by laser microdissection and reverse transcriptase PCR. Kiuru-Kuhlefelt et al13 used comparative genomic hybridization to analyze DNA copy number changes in the 17q and 22q regions and found an increase in copy number changes between dermatofibrosarcoma protuberans and fibrosarcomatous dermatofibrosarcoma protuberans, although the change was not statistically significant.
The results of our study support the hypothesis that gains in copy number of the COL1A1-PDGFB fusion gene may contribute to the fibrosarcomatous change in dermatofibrosarcoma protuberans in a subset of tumors since we identified six of 10 patients (60%) with fibrosarcomatous dermatofibrosarcoma protuberans that had statistically significant genomic gains of the COL1A1-PDGFB fusion gene in the fibrosarcomatous area over the dermatofibrosarcoma protuberans area. Our data also confirm the trend noted by Kiuru-Kuhlefelt, who used CGH to find a small increase in copy number changes of chromosome 17 and 22 material in a study set of similar size to our present study.13 Their data did not reach statistical significance and this finding not only reflects the difficulty CGH would have in detecting subtle changes of copy number but also that combining together cases that operate with and without gains of COL1A1-PDGFB in a nonpaired analysis may obscure individual tumor oncogenic mechanisms. The use of fluorescence in situ hybridization allows for individual cases and individual areas to be compared, and counting signals allows for fine differentiation in trends and the exclusion of normal cells such as blood vessels, adipocytes or inflammatory cells by morphologic examination during the counting process.
No cases in our study set showed average COL1A1-PDGFB fusion gene copy gains above 5.85 copies. This relatively small gain is in contrast to other many other oncogenes that can be amplified at a much higher level. For example, MYCN is usually amplified 50- to 100-fold in neuroblastoma and ERBB2 (HER2) can be amplified by up to 20 copies in breast carcinoma.19 One explanation of this finding is that relatively low copy numbers of COL1A1-PDGFB can already provide a potent oncogenic stimulus to drive tumor proliferation in dermatofibrosarcoma protuberans and modest changes in the fusion gene copy number has a major effect in the fibrosarcomatous transformation. However, higher levels can lead to toxic cell signaling that ultimately results in apoptosis, therefore, negatively selecting these clones in a similar manner to what was shown in MDM2 overexpressing mouse models.20
The observation that significant gains of the COL1A1-PDGFB fusion gene are not present in 40% of the study set implies that other oncogenic mechanisms must occur in the clonal evolution of fibrosarcomatous dermatofibrosarcoma protuberans. For example, both activating mutations in PDGFB or PDGFRB may lead to similar phenotypic effects. In addition to higher proliferation rates and gains of TP53 mutations, Kiuru-Kuhlefelt et al13 detected gains of whole or partial chromosomes 1 and 5 in their group of fibrosarcomatous dermatofibrosarcoma protuberans. Therefore, the amplification of the COL1A1-PDGFB fusion gene plays an important, but certainly not exclusive, role in the dedifferentiation of dermatofibrosarcoma protuberans into fibrosarcomatous dermatofibrosarcoma protuberans.
The genetic events that lead to the metastatic potential in fibrosarcomatous dermatofibrosarcoma protuberans are unknown. Previous retrospective clinicopathologic studies have shown an approximately 10% metastatic rate in fibrosarcomatous dermatofibrosarcoma protuberans, with lung and bone as common sites.2, 3 We analyzed a single case of fibrosarcomatous dermatofibrosarcoma protuberans that metastasized to a distant bone and the metastasis showed pure fibrosarcomatous morphology. The metastatic lesion possessed a slightly increased average COL1A1-PDGFB copy number gain over the fibrosarcomatous area, which in turn was similar to the average COL1A1-PDGFB copy number gain in the dermatofibrosarcoma protuberans area. This case demonstrates that the clonal selection for metastatic potential likely depends on multiple interrelated biologic factors and cannot be predicted by average COL1A1-PDGFB copy number gain. These data are consistent with data from epithelial tumors that show genetic instability quantitatively rises with tumor grade and aggressive behavior.21
The presence of 100% fibrosarcomatous morphology in the metastatic lesion with increased copy number gains of the COL1A1-PDGFB fusion gene highlights recent evidence that many superficial adult fibrosarcomas are indeed fibrosarcomatous dermatofibrosarcoma protuberans. In a case series by Sheng et al, four of six cases of superficial adult fibrosarcoma expressed the COL1A1-PDGFB fusion gene as detected by RT-PCR.22 Importantly, genetic analysis of clinically aggressive fibrosarcomas may become clinically warranted since the presence of the COL1A1-PDGFB fusion gene carries potential therapeutic implications. Inhibitors of PDGFB such as imatinib mesylate have been shown to have activity against primary and metastatic dermatofibrosarcoma protuberans and fibrosarcomatous dermatofibrosarcoma protuberans in small trials.23, 24, 25 In contrast, a small subset of dermatofibrosarcoma protuberans patients have shown transient or partial responses to imatinib.25, 26 The role of PDGFB activation in imatinib responsiveness has not been investigated, but it is possible that the subset of fibrosarcomatous dermatofibrosarcoma protuberans that operate with gains of COL1A1-PDGFB may be more susceptible to imatinib.24 However, our data also clearly indicate that additional genetic factors such as gains of TP53 mutations play an important role in tumor behavior.2 Therefore, investigation of other oncogenic mechanisms in the PDGFB-PDGFRB signaling pathway is necessary.
Our results showed that copy number of COL1A1-PDGFB fusion gene in the classic dermatofibrosarcoma protuberans areas does not seem to be a major predisposing mechanism for fibrosarcomatous transformation. There was basically no difference in COL1A1-PDGFB fusion gene copy gain in the dermatofibrosarcoma protuberans areas of fibrosarcomatous dermatofibrosarcoma protuberans and classic dermatofibrosarcoma protuberans. Therefore, while fibrosarcomatous areas may contain increased copies of the fusion gene in a large subset of tumors, the value of fusion gene copy numbers in the dermatofibrosarcoma protuberans areas is not predictive of clinical behavior. These data emphasize the role that multiple genetic events must play in clonal evolution of fibrosarcomatous change.
In summary, genomic gains of COL1A1-PDGFB fusion gene were identified in 60% of dermatofibrosarcoma protuberans that clonally evolve into fibrosarcomatous dermatofibrosarcoma protuberans. The remaining cases of fibrosarcomatous dermatofibrosarcoma protuberans do not show genomic gains of this fusion gene, which suggests other alternate oncogenic mechanisms must exist in this transformation. In addition, copy number of COL1A1-PDGFB fusion gene in the classic dermatofibrosarcoma protuberans areas does not seem to be a major predisposing mechanism for fibrosarcomatous transformation.
References
Gloster HM . Dermatofibrosarcoma protuberans. J Am Acad Dermatol 1996;35:355–374.
Abbott JJ, Oliveira AM, Nascimento AG . The prognostic significance of fibrosarcomatous transformation in dermatofibrosarcoma protuberans. Am J Surg Pathol 2006;30:436–443.
Mentzel T, Beham A, Katenkamp D, et al. Fibrosarcomatous (‘high-grade’) dermatofibrosarcoma protuberans: clinicopathologic and immunohistochemical study of a series of 41 cases with emphasis on prognostic significance [see comment]. Am J Surg Pathol 1998;22:576–587.
DuBay D, Cimmino V, Lowe L, et al. Low recurrence rate after surgery for dermatofibrosarcoma protuberans: a multidisciplinary approach from a single institution. Cancer 2004;100:1008–1016.
Gloster HM, Harris KR, Roenigk RK . A comparison between Mohs micrographic surgery and wide surgical excision for the treatment of dermatofibrosarcoma protuberans. J Am Acad Dermatol 1996;35:88–95.
Sirvent N, Maire G, Pedeutour F . Genetics of dermatofibrosarcoma protuberans family of tumors: from ring chromosomes to tyrosine kinase inhibitor treatment. Genes Chromosomes Cancer 2003;37:1–19.
Pedeutour F, Coindre JM, Nicolo G, et al. Ring chromosomes in dermatofibrosarcoma protuberans contain chromosome 17 sequences: fluorescence in situ hybridization. Cancer Genet Cytogenet 1993;67:149.
Pedeutour F, Simon MP, Minoletti F, et al. Translocation, t(17;22)(q22;q13), in dermatofibrosarcoma protuberans: a new tumor-associated chromosome rearrangement. Cytogenet Cell Genet 1996;72:171–174.
Simon MP, Pedeutour F, Sirvent N, et al. Deregulation of the platelet-derived growth factor B-chain gene via fusion with collagen gene COL1A1 in dermatofibrosarcoma protuberans and giant-cell fibroblastoma. Nat Genet 1997;15:95–98.
Shimizu A, O'Brien KP, Sjoblom T, et al. The dermatofibrosarcoma protuberans-associated collagen type Ialpha1/platelet-derived growth factor (PDGF) B-chain fusion gene generates a transforming protein that is processed to functional PDGF-BB. Cancer Res 1999;59:3719–3723.
Simon MP, Navarro M, Roux D, et al. Structural and functional analysis of a chimeric protein COL1A1-PDGFB generated by the translocation t(17;22)(q22;q13.1) in Dermatofibrosarcoma protuberans (DP). Oncogene 2001;20:2965–2975.
Wang J, Morimitsu Y, Okamoto S, et al. COL1A1-PDGFB fusion transcripts in fibrosarcomatous areas of six dermatofibrosarcomas protuberans. J Mol Diagn 2000;2:47–52.
Kiuru-Kuhlefelt S, El-Rifai W, Fanburg-Smith J, et al. Concomitant DNA copy number amplification at 17q and 22q in dermatofibrosarcoma protuberans. Cytogenet Cell Genet 2001;92:192–195.
Bowne WB, Antonescu CR, Leung DH, et al. Dermatofibrosarcoma protuberans: a clinicopathologic analysis of patients treated and followed at a single institution. Cancer 2000;88:2711–2720.
Hisaoka M, Okamoto S, Morimitsu Y, et al. Dermatofibrosarcoma protuberans with fibrosarcomatous areas. molecular abnormalities of the p53 pathway in fibrosarcomatous transformation of dermatofibrosarcoma protuberans. Virchows Arch 1998;433:323–329.
Takahira T, Oda Y, Tamiya S, et al. Microsatellite instability and p53 mutation associated with tumor progression in dermatofibrosarcoma protuberans. Hum Pathol 2004;35:240–245.
Sasaki M, Ishida T, Horiuchi H, et al. Dermatofibrosarcoma protuberans: an analysis of proliferative activity, DNA flow cytometry and p53 overexpression with emphasis on its progression. Pathol Int 1999;49:799–806.
Saeki H, Hoashi T, Tada Y, et al. Analysis of gene mutations in three cases of dermatofibrosarcoma protuberans (DFSP): ordinary DFSP, DFSP with fibrosarcomatous lesion (DFSP-FS) and lung metastasis of DFSP-FS. J Dermatol Sci 2003;33:161–167.
Schwab M . Oncogene amplification in solid tumors. Semin Cancer Biol 1999;9:319–325.
Jones SN, Hancock AR, Vogel H, et al. Overexpression of Mdm2 in mice reveals a p53-independent role for Mdm2 in tumorigenesis. Proc Natl Acad Sci 1998;95:15608–15612.
Ried T, Heselmeyer-Haddad K, Blegen H, et al. Genomic changes defining the genesis, progression, and malignancy potential in solid human tumors: a phenotype/genotype correlation. Genes Chromosomes Cancer 1999;25:195–204.
Sheng WQ, Hashimoto H, Okamoto S, et al. Expression of COL1A1-PDGFB fusion transcripts in superficial adult fibrosarcoma suggests a close relationship to dermatofibrosarcoma protuberans. J Pathol 2001;194:88–94.
McArthur GA, Demetri GD, van Oosterom A, et al. Molecular and clinical analysis of locally advanced dermatofibrosarcoma protuberans treated with imatinib: Imatinib Target Exploration Consortium Study B2225. J Clin Oncol 2005;23:866–873.
Labropoulos SV, Fletcher JA, Oliveira AM, et al. Sustained complete remission of metastatic dermatofibrosarcoma protuberans with imatinib mesylate. Anticancer Drugs 2005;16:461–466.
Rubin BP, Schuetze SM, Eary JF, et al. Molecular targeting of platelet-derived growth factor B by imatinib mesylate in a patient with metastatic dermatofibrosarcoma protuberans. J Clin Oncol 2002;20:3586–3591.
Maki RG, Awan RA, Dixon RH, et al. Differential sensitivity to imatinib of 2 patients with metastatic sarcoma arising from dermatofibrosarcoma protuberans. Int J Cancer 2002;100:623–626.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Abbott, J., Erickson-Johnson, M., Wang, X. et al. Gains of COL1A1-PDGFB genomic copies occur in fibrosarcomatous transformation of dermatofibrosarcoma protuberans. Mod Pathol 19, 1512–1518 (2006). https://doi.org/10.1038/modpathol.3800695
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.3800695
Keywords
This article is cited by
-
Coamplification of 12q15 and 12p13 and homozygous CDKN2A/2B deletion: synergistic role of fibrosarcomatous transformation in dermatofibrosarcoma protuberans with a cryptic COL1A1-PDGFB fusion
Virchows Archiv (2022)
-
PDGFB RNA in situ hybridization for the diagnosis of dermatofibrosarcoma protuberans
Modern Pathology (2021)
-
Cutaneous soft tissue tumors: how do we make sense of fibrous and “fibrohistiocytic” tumors with confusing names and similar appearances?
Modern Pathology (2020)
-
Soft Tissue Special Issue: Fibroblastic and Myofibroblastic Neoplasms of the Head and Neck
Head and Neck Pathology (2020)
-
Molecular testing for the clinical diagnosis of fibrolamellar carcinoma
Modern Pathology (2018)