Abstract
Image cytometric DNA ploidy analysis of endometrial carcinomas was performed to determine whether ploidy status and ploidy-related parameters like DNA index, percentage of cells exceeding 5c and 9c, correlate with histologic subtype. This is a prospective study of 391 patients with stage I endometrial carcinoma which included 331 (85%) endometrioid adenocarcinoma, 22 (6%) serous adenocarcinoma, 7 (2%) clear cell adenocarcinoma, 2 (0.5%) small cell carcinoma, 1 (0.3%) undifferentiated carcinoma, and 28 (7%) unclassifiable adenocarcinoma. Twenty-five percent of endometrioid adenocarcinomas were non-diploid. In contrast, all clear cell adenocarcinomas and 21/22 (95%) of serous adenocarcinomas were non-diploid. Hyperdiploidy (25 cases) was found only in endometrioid adenocarcinomas. Mean DNA index of the stemline in serous adenocarcinoma (1.72) and clear cell adenocarcinoma (1.81) was higher than in endometrioid adenocarcinoma (1.1). The difference in ploidy-related parameters between endometrioid adenocarcinoma and serous adenocarcinoma was highly significant (P<0.001). In addition, Grade 3 endometrioid adenocarcinoma showed significant difference in all ploidy-related parameters compared with grade 1 and grade 2 tumors (P<0.001). Our results show that DNA ploidy-related parameters may be valuable in subtyping histologically difficult cases of endometrial carcinomas.
Similar content being viewed by others
Main
DNA ploidy is known to be a strong prognostic parameter in endometrial carcinomas.1, 2, 3, 4, 5, 6, 7, 8, 9 In a few studies DNA ploidy results have been used as a predictive marker for treatment options.10 DNA ploidy-related parameters such as DNA index,8, 11 percentage of cells exceeding 5c (5c ER) also provide prognostic information.12
In the majority of DNA ploidy studies flow cytometry has been used on paraffin-embedded material13, 14, 15, 16 or frozen material.17, 18, 19 The advantage is the analysis of 20 000–50 000 nuclei. However, tumor cells cannot be separated from control cells in a sample, and the disturbance due to nuclear fragments may interfere with the evaluation of percentage of 5c and 9c exceeding nuclei especially in paraffin-embedded material. In some studies DNA histograms of a substantial number of cases were difficult to interpret.3 Moreover, small subpopulations of non-diploid cells may not be detected by flow cytometry.20
DNA-image cytometry has so far been a rather course method as only 100–200 nuclei have been measured.9, 14 The advantage of this method is that nuclear morphology can be evaluated. The DNA-image cytometry method presented in this study analyzing more than 1000 nuclei with evaluation of nuclear morphology has improved resolution. This also enables calculation of more precise percentage of nuclei exceeding 5c and 9c.
Histologic subtyping of endometrial carcinoma into endometrioid and non-endometrioid has been shown to predict survival.9, 13, 15 Serous adenocarcinoma and clear cell adenocarcinoma are aggressive tumors21 and are usually aneuploid.11 Grade 1 endometrioid adenocarcinoma has better survival21 and is usually diploid.13 The reviewed diagnosis has been reported to better correlate with survival compared to routine typing and grading.22 No comprehensive study comparing DNA ploidy with subtype and grade of endometrial carcinoma has been performed.23 In our study, we have compared reviewed subtype and grade, with high-resolution DNA image cytometric ploidy analysis on a large prospective, consecutive material of endometrial carcinomas from patients with stage I disease.
Materials and methods
This is a prospective study of endometrial carcinomas referred to Rikshospitalet-Radiumhospitalet Medical Center, Oslo, Norway between April 2003 and April 2005. During the 2 years, 399 specimens from patients with stage I disease have been analyzed. Representative areas (most atypical, best preserved, without necrosis and blood) were marked and processed for ploidy analysis. Eight cases (2%) with less than 300 nuclei analyzed were excluded from the study. Pathology reports were reviewed for age of the patients and type of specimen in which DNA ploidy was performed. Of the remaining 391 specimens, 165 (42%) analyses were performed on material from curettage specimen and 226 (58%) from hysterectomy specimens.
Histopathology
All H&E sections from paraffin-embedded tissue used for ploidy analyses were reviewed by two pathologists (VA and MP) for histological subtyping and grading according to the recommendations of World Health Organization (WHO) classification of tumors.24 Only tumors with a histological appearance which were in complete agreement with the WHO recommendation were classified as endometrioid adenocarcinoma, serous adenocarcinoma, clear cell adenocarcinoma, small cell carcinoma, and undifferentiated carcinoma. Tumors, which were showing features of adenocarcinoma but could not be classified according to WHO criteria, were diagnosed as adenocarcinoma unclassifiable. They included (1) tumors with papillary growth pattern but difficult to distinguish between serous adenocarcinoma and endometrioid adenocarcinoma (Figure 1d1 and g1) (2) tumors with clear cytoplasm, but difficult to distinguish between clear cell adenocarcinoma and secretory variant of endometrioid adenocarcinoma (Figure 1i1) (3) tumors showing glandular differentiation and composed of two types of cells lining the glands, one with elongated cells with vesicular nuclei and another with slender cells with hyperchromatic nuclei lying perpendicular to the basement membrane (Figure 1e1) (4) tumors showing areas with highly pleomorphic nuclei (Figure 1j1).
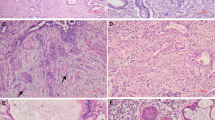
Histology and DNA histogram of endometrial carcinomas. (a1, a2) Endometrioid adenocarcinoma grade 1 with diploid histogram. (b1, b2) Endometrioid adenocarcinoma with hyperdiploid DNA histogram. (c1, c2) Endometrioid adenocarcinoma grade 3 showing aneuploid peak at DNA index 1.2 in DNA histogram. (d1, d2) Adenocarcinoma unclassifiable with diploid DNA histogram, possibly a case of endometrioid adenocarcinoma. (e1, e2) Adenocarcinoma unclassifiable demonstrating vesicular and slender nuclei with diploid DNA histogram, possibly a variant of endometrioid adenocarcinoma. (f1, f2) Serous adenocarcinoma with aneuploid peak at DNA index 1.7. (g1, g2) Adenocarcinoma unclassifiable with aneuploid peak at DNA index 1.7, possibly a case of serous adenocarcinoma. (h1, h2) Clear cell adenocarcinoma with tetraploid DNA histogram. (i1, i2) Adenocarcinoma unclassifiable showing clear cytoplasm and tetraploid histogram, possibly a case of clear cell adenocarcinoma. (j1, j2) Adenocarcinoma unclassifiable showing markedly pleomorphic nuclei with aneuploid DNA histogram.
Tumors with ≤5% solid area were graded as grade 1, 6–50% solid area as grade 2, and >50% solid area as grade 3. Squamous component was excluded from grading and in tumors with bizarre atypia, grade was raised by one (from 1 to 2 or 2 to 3). Grading was only done in endometrioid adenocarcinoma. Serous adenocarcinoma, clear cell adenocarcinoma, small cell carcinoma, undifferentiated carcinoma, and adenocarcinoma unclassifiable were not graded.
DNA Image Cytometry
Monolayer preparation and Feulgen's staining
Two or more 50-μm sections, containing more than 90% representative tumor tissue, were cut from the paraffin-embedded tissue blocks. A 5-μm section, cut and stained with hematoxylin and eosin (H&E), was used for control and for reviewing the diagnosis. The sections were deparaffinized in xylene, rehydrated through decreasing alcohol gradient, and washed twice in phosphate-buffered saline (PBS). The sections were incubated at 37°C at 200 rpm for 1 h with 0.5 mg/ml protease (Sigma P8038) to disaggregate the cells. Cold PBS was added and the tubes were placed in ice bath to stop enzymatic digestion. The cell suspension was filtered through a 60-μm mesh nylon filter and centrifuged at 1000 rpm (Sigma 3K-1) for 10 min. After discarding the supernatant, the pellet was resuspended in PBS. A volume of 100 μl of the solution was cytospined at 600 rpm for 5 min to prepare a monolayer on a poly-L-lysin coated slide. Cell density on the slide was checked under microscope and additional spins were performed if correction of the density was required. The monolayer preparations were air-dried and fixed overnight in 4% formaldehyde.
The monolayer preparations were stained using Feulgen's stain. After washing the slides in distilled water, they were kept in 5 M HCl for 60 min at room temperature for hydrolysis. Afterwards slides were rinsed and stained with Schiff's solution for 2 h in the dark. The preparations were then washed in water and left in freshly made solution of 0.5% Sodium disulphite in 0.05 M HCl. Finally, the slides were washed in running tap water, dehydrated in increasing alcohol gradient, cleared in Xylene, and coverslipped.
Measurement of DNA content
Nuclei stained with Feulgen's stain were used for measuring DNA content with the use of Fairfield DNA Ploidy system (Fairfield Imaging, Nottingham, UK). It consisted of Zeiss Axioplan microscope (Zeiss, Jena, Germany) equipped with a 40/0.75 objective lens, a 546 nm green filter and a black and white high-resolution digital camera (model C4742-95, Hamamatsu Photonics K.K., Hamamatsu, Japan). Optical density and nuclear area were measured and integrated optical density of each nucleus was calculated. Background optical density was corrected for each nucleus.
At least 1500 nuclei were automatically scanned and images were collected into galleries: nuclei of interest for measurement; lymphocytes, plasma cells and fibroblast as reference cells. The galleries were edited to discard cut, overlapped and pyknotic nuclei. Cases with less than 300 nuclei of interest were excluded from the study. Histogram Draftsman 1.4 (Fairfield Imaging Ltd) was used to create histogram from integrated optical density of nuclear images. The reference-cell nuclei were used as an internal diploid control.
Histograms were classified according to the below-mentioned criteria. Ploidy-related parameters as DNA index (DI) and percentages of cells exceeding 5c (5c ER) and 9c (9c ER) were noted.
Criteria for the classification of DNA ploidy
A tumor was classified as diploid if only one G0/G1 peak (2c) was present, the number of nuclei at G2 peak (4c) did not exceed 10% of the total number of nuclei and the number of nuclei with DNA content more than 5c did not exceed 1%. A tumor was defined as tetraploid if a 4c peak (DI 1.9–2.1) was present (the number of nuclei at 4c peak is more than the S-phase fraction and more than 10% of the total nuclei), the number of nuclei at G2 peak (8c) did not exceed 10% of the total number of nuclei and the number of nuclei with DNA content more than 9c did not exceed 1%. A tumor was defined as polyploid when more than 10% of total number of nuclei was present at 8c peak and/or number of nuclei with DNA content more than 9c exceeded 1%. A tumor was defined as aneuploid when non-euploid peak(s) were present or the number of nuclei with a DNA content exceeding 5c, not representing euploid populations, exceeded 1%.25 A tumor was classified as hyperdiploid if an aneuploid peak was demonstrated with DNA index 1.06–1.10.
Statistical Analysis
Statistical analysis was done with SPSS (Statistical Package for the Social Sciences) version 12.0.1 (SPSS Inc, Chicago, IL). Analysis of variance was performed to explore the impact of histologic subtype and grade to ploidy-related parameters. Histologic groups were compared with each other by using Tukey HSD test. Independent sample t-test and χ2-test were used to compare endometrioid and non-endometrioid group.
Results
Age of the patients ranged between 29 and 96 years (mean, 65). Ploidy status, histologic subtypes and grades of the 391 endometrial carcinomas are given in Table 1. Mean number of nuclei analyzed was 1145 (range 388–1714). Mean coefficient of variation of stemline was 3.2% (range 0.87–8.01).
Endometrioid Adenocarcinoma
In all, 175 (45%) of the 331 endometrioid adenocarcinomas were grade 1, 118 (30%) grade 2 and 38 (10%) grade 3. In four cases of endometrioid adenocarcinoma bizarre atypia was observed and grades were increased by 1. A total of 249 cases (75 %) were diploid (Figure 1a1 and a2) and 38 cases (12%) were aneuploid (Figure 1c1 and c2). More than three-fourth of Grade 1 and 2, and only half of grade 3 endometrioid carcinomas were diploid (Table 1). Hyperdiploidy was only found in endometrioid adenocarcinoma (25 cases) and mainly in grade 1 and 2 tumors (23 cases, Figure 1b1 and b2).
Serous Adenocarcinoma
Seventeen (77%) of the 22 serous adenocarcinomas were aneuploid and 4 were tetraploid. Fifteen tumors (68%) had a DNA index between 1.6 and 1.89 (Figure 1f1 and f2). Highest mean 5c ER, 9c ER were found in serous adenocarcinoma (Table 1). One of the tumors was diploid.
Clear Cell Adenocarcinoma
All the seven cases of clear cell adenocarcinoma were non-diploid. Four (57%) of the cases were tetraploid (Figure 1h1 and h2) and three (43%) aneuploid. Highest mean DNA index (1.81) was observed in clear cell adenocarcinoma (Table 1).
Adenocarcinoma Unclassifiable
Twenty-eight cases were diagnosed as adenocarcinoma unclassifiable, which consists of four different groups. (1) Thirteen cases with papillary growth pattern which were difficult to distinguish between serous adenocarcinoma and endometrioid adenocarcinoma (Figure 1d1 and g1); ploidy analysis showed four diploid cases, four tetraploid, and five aneuploid with DNA index 1.13, 1.14, 1.66, 1.67, and 1.69. (2) Six cases with clear cytoplasm were difficult to distinguish between clear cell adenocarcinoma and endometrioid adenocarcinoma of the secretory variant (Figure 1i1). In this group two cases were diploid, two were tetraploid and two were aneuploid with DNA index 1.83 and 2.15. (3) One case with highly pleomorphic nuclei (Figure 1j1) showed aneuploid histogram with DNA index 1.37. (4) Eight cases showed glandular differentiation as endometrioid adenocarcinoma but two types of cells were lining the glands. One was elongated cells with vesicular nuclei and another, more slender cells with hyperchromatic nuclei lying perpendicular to the basement membrane (Figure 1e1). Of the eight cases, seven were diploid and one tetraploid. Out of all the cases of adenocarcinoma unclassifiable, 13 (46%) were diploid, 8 (29%) were aneuploid and 7 (25%) were tetraploid.
Small Cell Carcinoma and Undifferentiated Carcinoma
The two cases of small cell carcinoma showed diploid histogram. Tetraploid histogram was seen in a case of undifferentiated carcinoma.
Statistical Analysis
A one-way between-groups analysis of variance was conducted between histological diagnoses and DNA index, 5cER and 9cER. There were significant differences at P<0.001 in all the parameters. Post-hoc comparison using Tukey HSD test to evaluate the difference of individual carcinoma showed that means of the DNA index, 5cER and 9cER of endometrioid adenocarcinoma were significantly different from serous adenocarcinoma and clear cell adenocarcinoma (Table 2). However, no significant difference was seen between serous adenocarcinoma and clear cell adenocarcinoma. Similar analysis was also conducted between grades of endometrioid carcinoma which showed significant differences at P<0.001 in all parameters. In post-hoc comparison grade 1 and grade 2 endometrioid adenocarcinomas did not differ significantly; however, both groups differed significantly from grade 3 endometrioid adenocarcinoma in all ploidy-related parameters (Table 2).
Statistical analysis was also performed by dividing histological subtypes into type 1 (endometrioid adenocarcinoma) and type 2 carcinomas (serous adenocarcinoma and clear cell adenocarcinoma), and ploidy results into diploid and non-diploid. Chi-square test showed that type 1 carcinoma significantly differed from type 2 carcinoma (P<0.001) regarding diploid vs non-diploid. An independent-samples t-test was conducted to compare DNA index, 5cER and 9cER values for type 1 and type 2 endometrial carcinomas. There was significant difference in DNA index (P<0.001), 5c ER (P<0.001), and 9cER (P=0.002).
Discussion
In the current study we found that ploidy status correlates with histological subtypes of endometrial carcinoma. Most of the endometrioid adenocarcinomas were diploid (Figure 1a1 and a2). Almost all (95%) of the serous adenocarcinomas and all clear cell adenocarcinomas were non-diploid. Correlation of ploidy with endometrioid and non-endometrioid subtypes have been reported previously.13 In this study we have confirmed this and further demonstrated the relation of ploidy and ploidy-related parameters with individual histologic subtypes.
The present study differs from former ones in the use of high-resolution DNA ploidy histogram in which a larger number of nuclei are analyzed with simultaneous morphological observation of all nuclei. Previously 100–200 nuclei were analyzed.9, 14 In contrast, mean number of nuclei analyzed in our study was 1145. We used paraffin-embedded material for DNA cytometry and produced high-resolution histogram. This is also in contrast to the studies using DNA flow cytometry of paraffin-embedded material where as many as 55% (162/293) histograms have been reported as technically suboptimal.3 The high resolution is demonstrated by very low coefficient of variation (CV, mean=3.2%) of main stem line peak, which is less than CV of flow cytometric analysis on paraffin-embedded material.11 A previous study in our laboratory demonstrated that image cytometry was superior to flow cytometry in detecting aneuploid population.20 We, therefore, use only image cytometry DNA ploidy analysis in our laboratory.
We have classified ploidy histograms into diploid, hyperdiploid, aneuploid, tetraploid and polyploid. Many authors have used a classification with diploid and aneuploid13, 14 possibly due to the application of flow cytometry method and to ease the statistical analysis. In some studies tetraploid and diploid have been analyzed as a single group.9, 26 In our opinion, tetraploid tumors compose a distinct entity and should not be grouped as diploid as 21% of grade 3 endometrioid adenocarcinomas and more than half of the clear cell adenocarcinomas show tetraploidy. Other authors15, 16 have grouped tetraploid and aneuploid together and have shown that they differ significantly from the diploid group concerning prognosis.
Endometrial carcinoma has also been classified as type 1 (estrogen dependent) and as type 2 (estrogen independent). In this study Type 1 and type 2 carcinomas differed significantly from each other with respect to the ploidy-related parameters: DNA index, 5cER and 9cER. In addition, the two subtypes of estrogen independent carcinoma (type 2), serous adenocarcinoma and clear cell adenocarcinoma, did not differ from each other concerning these parameters (P>0.05). This finding also supports the logic of including these carcinomas in type 2.
Endometrioid adenocarcinoma, being 85% of all endometrial carcinomas, is not a homogenous group. There are many subtypes, and tumors are further subdivided by three grades according to the glandular differentiation. We showed that grade 3 carcinoma differed from grade 1 and 2 (P<0.001) in all ploidy-related parameters, but grade 1 and grade 2 did not differ significantly (P>0.05). The difference between grades has also been demonstrated by comparative genomic hybridization findings which showed gradual increase in the number of gains and losses from grade 1 to grade 3 endometrioid carcinoma.27
In our study endometrioid adenocarcinomas were mainly diploid, whereas serous and clear cell adenocarcinomas were mainly aneuploid. Serous and clear cell adenocarcinomas showed higher 5c ER and 9c ER than endometrioid adenocarcinomas. The difference between endometrioid carcinomas and non-endometrioid carcinomas has also been revealed by findings of comparative genomic hybridization which demonstrated that endometrioid adenocarcinomas have different gains and losses than non-endometrioid carcinomas, whereas serous adenocarcinomas and clear cell adenocarcinomas have similar gains and losses.27
We have observed that a subtype often demonstrates DNA index within a limited range (Figure 2). Most of serous adenocarcinomas (68%) had their stemline between DNA indexes 1.6–1.89. This is likely due to distinctive types of genetic instability and molecular alterations.23 In addition, more than half of the clear cell adenocarcinomas were tetraploid. Hyperdiploidy was only seen in endometrioid adenocarcinoma in our study (Figure 1b1 and b2). This might be due to a single additional chromosome, mainly found in well differentiated endometrioid adenocarcinomas with favorable prognosis.28 These findings indicate a solid biological background for modern histopathological subtyping of endometrial carcinomas.
Distribution of DNA indexes of endometrial carcinoma in percent. (a) Most of the endometrioid adenocarcinomas were diploid, some were hyperdiploid and tetraploid. (b) Most of the serous adenocarcinomas had DNA index between 1.7 and 2. (c) Clear cell adenocarcinomas were mostly tetraploid. (d) Adenocarcinoma unclassifiable had a heterogenous DNA index.
In two cases of small cell carcinoma, despite an aggressive tumor type, diploid histogram is seen. This is peculiar but the number is too small to draw any conclusion. In this series we identified only one case of undifferentiated carcinoma with tetraploidy.
Following the WHO classification criteria for subtyping endometrial carcinoma, we found that 7% of the total tumors were not fulfilling the criteria and considered them as adenocarcinoma unclassifiable. The ploidy results of these tumors were heterogenous indicating a mixture of subtypes (Figure 2d). The three unclassifiable tumors, with papillary processes showing aneuploidy with DNA index between 1.6 and 1.89, are most likely serous adenocarcinoma rather than endometrial adenocarcinoma (Figure 1g1 and g2). Similarly six unclassifiable tumors showing diploidy and aneuploidy with DNA index between 1 and 1.6 are likely to represent endometrioid carcinoma (Figure 1d1 and d2). Two tumors with clear cytoplasm showing tetraploidy and two aneuploid tumors with DNA index 1.83 and 2.15 are most likely to be clear cell adenocarcinoma (Figure 1i1 and i2). Similarly, two diploid tumors are possibly secretory variant of endometrioid carcinoma. Seven out of eight cases having two types of cells lining the glands showed diploid histogram. This group showed ploidy results similar to endometrioid carcinoma. Therefore, this group possibly is a variant of endometrioid adenocarcinoma. These data have to be correlated to the clinical outcome of the patients.
In conclusion, DNA ploidy status and DNA ploidy-related parameters correlate with histopathological subtypes of endometrial carcinoma and may be valuable for subtyping the difficult cases.
References
Lindahl B, Alm P, Fernö M, et al. Prognostic value of flow cytometrical DNA measurements in stage I–II endometrial carcinoma: correlations with steroid receptor concentration, tumor myometrial invasion, and degree of differentiation. Anticancer Res 1987;7:791–797.
Iversen OE . Flow cytometric deoxyribonucleic acid index: a prognostic factor in endometrial carcinoma. Am J Obstet Gynecol 1986;155:770–776.
Zaino RJ, Davis AT, Ohlsson-Wilhelm BM, et al. DNA content is an independent prognostic indicator in endometrial adenocarcinoma. A Gynecologic Oncology Group study. Int J Gynecol Pathol 1998;17:312–319.
Mangili G, De Marzi P, Vigano R, et al. Identification of high-risk patients with endometrial carcinoma. Prognostic assessment of endometrial cancer. Eur J Gynaecol Oncol 2002;23:216–220.
Larson DM, Berg R, Shaw G, et al. Prognostic significance of DNA ploidy in endometrial cancer. Gynecol Oncol 1999;74:356–360.
Britton LC, Wilson TO, Gaffey TA, et al. DNA ploidy in endometrial carcinoma: major objective prognostic factor. Mayo Clin Proc 1990;65:643–650.
Wimberger P, Hillemanns P, Kapsner T, et al. Evaluation of prognostic factors following flow-cytometric DNA analysis after cytokeratin labelling: II. Cervical and endometrial cancer. Anal Cell Pathol 2002;24:147–158.
Lindahl B, Ranstam J, Willén R . Five year survival rate in endometrial carcinoma stages I–II: influence of degree of tumour differentiation, age, myometrial invasion and DNA content. Br J Obstet Gynaecol 1994;101:621–625.
Lundgren C, Auer G, Frankendal B, et al. Nuclear DNA content, proliferative activity, and p53 expression related to clinical and histopathologic features in endometrial carcinoma. Int J Gynecol Cancer 2002;12:110–118.
Högberg T, Fredstorp-Lidebring M, Alm P, et al. A prospective population-based management program including primary surgery and postoperative risk assessment by means of DNA ploidy and histopathology. Adjuvant radiotherapy is not necessary for the majority of patients with FIGO stage I–II endometrial cancer. Int J Gynecol Cancer 2004;14:437–450.
Newbury R, Schuerch C, Goodspeed N, et al. DNA content as a prognostic factor in endometrial carcinoma. Obstet Gynecol 1990;76:251–257.
Strang P, Stenkvist B, Bergström R, et al. Flow cytometry and interactive image cytometry in endometrial carcinoma. A comparative and prognostic study. Anticancer Res 1991;11:783–788.
Osmanağaoğlu MA, Kadioğlu S, Osmanağ S, et al. The relationship between mutant p53 gene, DNA contents and conventional clinicopathological prognostic variables in cases with endometrial carcinoma. Eur J Gynaecol Oncol 2005;26:64–70.
Terada K, Mattson D, Goo D, et al. DNA aneuploidy is associated with increased mortality for stage I endometrial cancer. Gynecol Oncol 2004;95:483–487.
Mariani A, Sebo TJ, Webb MJ, et al. Molecular and histopathologic predictors of distant failure in endometrial cancer. Cancer Detect Prev 2003;27:434–441.
Jhala DN, Atkinson BF, Balsara GR, et al. Role of DNA ploidy analysis in endometrial adenocarcinoma. Ann Diagn Pathol 2001;5:267–273.
Santala M, Talvensaari-Mattila A . DNA ploidy is an independent prognostic indicator of overall survival in stage I endometrial endometrioid carcinoma. Anticancer Res 2003;23:5191–5196.
Sorbe B, Risberg B . Prognostic importance of the nuclear proteins p53 and Rb in conjunction with DNA, nuclear morphometry and grading in endometrial carcinoma. Int J Gynecol Cancer 1997;7:34–41.
Koul A, Willen R, Bendahl PO, et al. Distinct sets of gene alterations in endometrial carcinoma implicate alternate modes of tumorigenesis. Cancer 2002;94:2369–2379.
Kaern J, Wetteland J, Tropé CG, et al. Comparison between flow cytometry and image cytometry in ploidy distribution assessments in gynecologic cancer. Cytometry 1992;13:314–321.
Abeler VM, Kjørstad KE, Berle E . Carcinoma of the endometrium in Norway: a histopathological and prognostic survey of a total population. Int J Gynecol Cancer 1992;2:9–22.
Baak JP, Snijders WP, van Diest PJ, et al. Confirmation of the prognostic value of the ECPI-1 score (myometrial invasion, DNA-ploidy and mean shortest nuclear axis) in FIGO stage I endometrial cancer patients with long follow-up. Int J Gynecol Cancer 1995;5:112–116.
Lax SF . Molecular genetic pathways in various types of endometrial carcinoma: from a phenotypical to a molecular-based classification. Virchows Arch 2004;444:213–223.
Silverberg SG, Kurman RJ, Nogales F, et al. Tumours of uterine corpus. In: Tavassoli FA, Devilee P (eds). World Health Organization Classification of Tumours: Pathology and Genetics. Tumours of the Breast and Female Genital Organs. IARC: Lyon, France, 2003, pp 218–232.
Kristensen GB, Kildal W, Abeler VM, et al. Large-scale genomic instability predicts long-term outcome for women with invasive stage I ovarian cancer. Ann Oncol 2003;14:1494–1500.
Baak JP, Snijders W, van Diermen B, et al. Prospective multicenter validation confirms the prognostic superiority of the endometrial carcinoma prognostic index in international Federation of gynecology and obstetrics stage 1 and 2 endometrial carcinoma. J Clin Oncol 2003;21:4214–4221.
Micci F, Teixeira MR, Haugom L, et al. Genomic aberrations in carcinomas of the uterine corpus. Genes Chromosomes Cancer 2004;40:229–246.
Atkin NB . Frequency of hyperdiploid chromosome complements in endometrioid tumors of the endometrium whereas similar tumors in the ovary tend to show hypodiploidy: a significant difference that may not be distinguishable by flow cytometry of DNA content. Cytogenet Genome Res 2002;97:39–42.
Acknowledgements
We thank Mrs Signe Eastgate and Mrs Erika Thorbjørnsen for their skillful technical assistance. This study was supported by The Norwegian Radium Hospital Research Foundation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Pradhan, M., Abeler, V., Danielsen, H. et al. Image cytometry DNA ploidy correlates with histological subtypes in endometrial carcinomas. Mod Pathol 19, 1227–1235 (2006). https://doi.org/10.1038/modpathol.3800641
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.3800641
Keywords
This article is cited by
-
Predicting malignant progression in clinically high-risk lesions by DNA ploidy analysis and dysplasia grading
Scientific Reports (2018)
-
Revisiting tumour aneuploidy — the place of ploidy assessment in the molecular era
Nature Reviews Clinical Oncology (2016)
-
DNA ploidy in curettage specimens identifies high-risk patients and lymph node metastasis in endometrial cancer
British Journal of Cancer (2015)
-
Cooperation between p53 and the telomere-protecting shelterin component Pot1a in endometrial carcinogenesis
Oncogene (2013)
-
DNA ploidy may be a prognostic marker in stage I and II serous adenocarcinoma of the endometrium
Virchows Archiv (2012)