Abstract
Clear cell sarcoma of soft parts is a rare soft tissue malignancy that shows phenotypic overlap with cutaneous melanoma but can be distinguished by the presence of a t(12;22) translocation. Microsatellite instability (MSI), a variation in the lengths of short repeat DNA segments in the genome, has been implicated in melanoma tumorigenesis, but is rare or absent in clear cell sarcoma. Defects in the mismatch repair (MMR) enzyme complex correlate with MSI in some tumor types, allowing the use of immunohistochemistry for the MMR proteins hMLH1 and hMSH2 to predict the presence of MSI. To determine if the association between MMR defects and MSI extends to clear cell sarcoma, we compared a group of nine clear cell sarcomas to 11 metastatic melanomas on the basis of MSI and the expression of MMR proteins. MSI was studied using fluorescence-based multiplexed PCR of five loci. Immunohistochemistry was evaluated on formalin-fixed paraffin-embedded tissue for hMLH1, hMHS2 and hMSH6. MSI was present in only 1/9 (11%) clear cell sarcoma case and in 8/11 (73%) melanoma cases. Immunostaining for hMLH1 and hMSH2 was preserved in all the clear cell sarcomas but loss of immunostaining for one or both proteins was seen in 6/11 melanomas (55%). hMSH6 was detected in 7/9 (78%) clear cell sarcomas and 10/11 (91%) of melanomas. Clear cell sarcoma and metastatic melanoma differed significantly with respect to the presence of MSI (P=0.010) and staining for hMLH1 and/or hMSH2 (P=0.014) but not hMSH6 (P=0.57). Mismatch repair, and consequently genomic instability may contribute to tumorigenesis in melanoma but not clear cell sarcoma. Immunostaining for hMLH1 and hMSH2 and MSI analysis may be helpful in the differential diagnosis of large soft tissue or visceral malignancies with melanocytic differentiation.
Similar content being viewed by others
Main
Clear cell sarcoma is a rare soft tissue malignancy primarily affecting the deep soft tissues of the lower extremities in young adults.1 Clear cell sarcoma demonstrates immunohistochemical and ultrastructural evidence of melanocytic differentiation,2, 3, 4, 5, 6, 7 but is characterized by a t(12;22)(q13;q12) translocation. The resulting fusion between the EWS and ATF1 genes has not been observed in melanoma, although it may not be exclusive to clear cell sarcoma.8 Clear cell sarcoma and metastatic melanoma are both associated with poor prognosis. Nevertheless, the 5-year survival rate of clear cell sarcoma may be superior to metastatic melanoma,5, 9 thus conferring clinical impact to an accurate diagnosis when the differential includes these entities. The distinction between clear cell sarcoma and a soft tissue metastasis from a cutaneous or mucosal melanoma can usually be made based on clinical parameters. However, in the case of a metastasis from an occult primary melanoma or a clear cell sarcoma in an unexpected location, the distinction may pose a diagnostic dilemma that can be resolved by molecular methods.10, 11, 12, 13, 14 The similarity in immunophenotype has lead to few immunohistochemical methods that differentiate between the two malignancies.15
Despite overlapping phenotypic features, little is known about the histogenic relationship between clear cell sarcoma and melanoma. In general, metastatic melanoma demonstrates both the chromosomal instability pathway, characterized by complex karyotypic abnormalities,16, 17, 18, 19 and the mutator pathway characterized by microsatellite instability (MSI).20, 21, 22, 23 Interestingly, clear cell sarcoma appears to be microsatellite stable, at least in the few cases thus far studied.24 MSI, the alteration in the length of microsatellite DNA, has been most well studied in carcinoma, but has also been identified in some sarcomas and melanoma.25, 26, 27, 28, 29 In many examples, MSI appears to be related to a loss of function of the mismatch repair (MMR) enzyme system.30 Defects in MMR affect the rate of spontaneous mutations leading to tumorigenesis and are markers of genetic instability.20 Defective, decreased or absent expression of three components of the MMR system, hMLH1, hMSH2 and hMSH6, is most commonly implicated in germline mutations in the syndrome of hereditary non-polyposis colon cancer (HNPCC).31, 32, 33 In sporadic tumors, immunohistochemical stains for hMLH1 and hMSH2 have been useful to measure reduced expression of these proteins and thus predict MSI.34, 35 The significance of the loss of hMSH6 expression in sporadic tumors is less well established.36, 37
A growing body of evidence also supports a role for MSI and defective MMR protein expression in sarcoma and melanoma tumorigenesis.24, 27, 29, 38, 39, 40 Reduced expression of hMSH2 has been associated with a worse prognosis in soft tissue sarcomas as a group,41 while absent hMLH1 and hMSH2 expression has been demonstrated in melanomas and correlates with tumor progression.42, 43, 44 Inactivation of hMLH1 or hMSH2 may correlate with MSI in some sarcoma types28 but this association has not been established for clear cell sarcoma. If MMR protein expression is preserved in clear cell sarcoma, as might be predicted from the absence of MSI previously reported24 but not metastatic melanoma, these markers may prove of diagnostic utility in ambiguous cases.
The purpose of this study was to examine the difference in MSI and MMR protein expression between clear cell sarcoma and soft tissue or visceral melanoma metastases. We employed fluorescence-based multiplex PCR to determine the presence of MSI in nine well-characterized clear cell sarcomas and 11 metastatic melanomas in which clear cell sarcoma may be considered in the differential diagnosis. Furthermore, we examined the expression of three MMR proteins, hMLH1, hMSH2 and hMSH6, by immunohistochemistry to compare the two groups of tumors.
Methods
Selection of Cases
Nine patients with clear cell sarcoma were identified from the Department of Pathology files of University of California, San Francisco based on light microscopy, immunophenotype and ultrastructure. We previously reported this series of tumors and confirmed the presence of the EWS-ATF1 fusion transcript by reverse transcriptase-PCR.15 The comparison group of eleven metastatic melanomas, also previously reported, consisted of patients with documented cutaneous primaries and lacked EWS-ATF1 by RT-PCR.15 None of the patients in either group met clinical criteria for hereditary non-polyposis colon cancer to suggest germ line mutations of MMR proteins.45
Microsatellite Instability
Appropriate areas of normal tissue and tumor were selected from H&E stained slides. The corresponding tissue (approximately 0.5 cm2) was microdissected from 10 μm paraffin sections on unbaked, unstained, glass slides. In all cases, normal and tumor tissue was obtained from the same slide. DNA was extracted using the DNeasy kit (Qiagen, Valencia, CA, USA) according to the manufacturer's directions. Each PCR reaction contained 5 μl of DNA template, 0.4 μM of each fluorescently labeled primer for one of five established microsatellite markers (two mononucleotide repeats: BAT25, BAT26; three dinucleotide repeats D5S346, D17S250 and D2S123),46, 47 and 9 μl of True Allele PCR Premix (Applied Biosystems, Foster City, CA, USA). The primer sequences for the microsatellite markers can be obtained from the Genome Database (www.gdb.org). The amplification profile was denaturation at 94°C for 30 s, annealing at 55°C for 45 s, extension at 72°C for 1 min over 40 cycles followed by a final extension step at 72°C for 10 min. The products were resolved using an automated sequencer (ABI Prism model 3700 Genetic Analyzer, Applied Biosystems, Foster City, CA, USA) using ROX-labeled molecular weight markers and the GeneScan software package (Applied Biosystems, Foster City, CA, USA). Instability at a given locus was defined by the presence of novel peaks, present in tumor DNA, but absent in normal DNA. Using criteria established for colorectal carcinoma, tumors that demonstrated instability of ≥2 informative loci were classified as high microsatellite instability (MSI-H) and those with instability at only 1 locus were classified as low instability (MSI-L).46 Loss of heterozygosity (LOH) was recorded when the ratio of peak heights of two normal alleles to peak heights of two tumor alleles was >2.0 or <0.5.48
Immunohistochemistry
Immunohistochemical analysis was performed using standard techniques.49 Briefly, 4 μm paraffin-embedded sections were heated, de-paraffinized, heated in citrate buffer, blocked for endogenous peroxidase, avidin and biotin, incubated with antibodies (Table 1), washed and developed using the LSAB kit (DAKO, Carpinteria, CA, USA). Colon adenocarcinomas with previously documented loss of nuclear hMSH1 and/or hMLH2 and normal colon were used as controls. Slides were scored by both JG and AEH who were blinded to the diagnosis and MSI results. In the area dissected for MSI analysis, staining was scored for the presence of nuclear hMLH1, hMSH2 or hMSH6 as negative if there was a complete absence of nuclear staining in tumor cells.28, 29, 50 Cases with either focal or diffuse staining were interpreted as positive. Nuclear staining in lymphocytes or adjacent normal tissue served as internal positive controls in negative cases.
Statistical Analysis
The relationship between variables was tested by the Fisher's exact test. P-values <0.05 were considered statistically significant.
Results
Clinical Findings
Clinicopathologic characteristics of the clear cell sarcoma and melanoma patients in this study are summarized in Table 2 and have been previously reported.15 The clear cell sarcoma patients ranged from 19 to 72 years (average, 44.8 years) and five of nine were male. Five patients presented with a primary lesion in the lower extremity (foot, leg or groin) while the others had tumors in the pelvis, axilla and arm (two). One patient (C8) also had a metastasis available for study. Tumor size in patients with clear cell sarcoma ranged from 2 to 9 cm. Of the 11 patients with metastatic melanoma, seven were male and ages ranged from 29 to 84 years (average, 59.9 years). Sites of tumor presentation in these patients were groin (three), lung (three), axilla (two), neck, adrenal gland and bowel. Melanoma tumor size ranged from 1 to 13 cm.
Microsatellite Instability
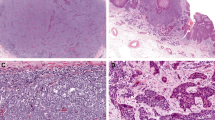
Microsatellite instability (MSI) was evaluated at five loci: BAT25, BAT26, D5S346, D17S250 and D2S123. Representative fragment patterns are shown in Figure 1 and the results are summarized in Table 2. Of the nine clear cell sarcomas, instability was observed in only one tumor (11%), at a single locus D5S346 (MSI-L). The remaining tumors showed neither MSI nor LOH. A lymph node metastasis from clear cell sarcoma patient C8 was also analyzed and showed identical results the primary tumor (data not shown). MSI was much more common in the metastatic melanoma cases: eight of 11 (73%) cases showed instability of at least one locus. More specifically, instability at one locus (MSI-L) was observed in three (27%) and instability at two or more loci (MSI-H) in five (46%) cases. The mononucleotide repeat BAT25, showed microsatellite stability in all melanomas but LOH at BAT25 was observed in two cases.
Representative microsatellite instability results in clear cell sarcoma and melanoma. Each figure (a–c) shows microsatellite data for DNA extracted from normal tissue (upper panel) and tumor (lower panel). Clear cell sarcoma C9 demonstrates stability at all five loci (a). Melanoma M4 shows instability at D17S250 and D2S123 and loss of heterozygosity at D5S346 (b). Melanoma M10 demonstrates stability at all five loci (c). The relative locations of the markers are indicated at the top of the figure as well by color as follows: D5S346 and D17S250 green; BAT25 and D2S123 blue; BAT26 black; molecular weight markers red.
Immunohistochemical Analysis of MMR Proteins
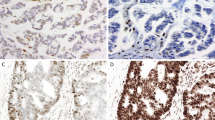
Representative immunohistochemical data are illustrated in Figure 2 and the results summarized in Table 2. The intensity of staining varied for these markers, with hMLH1 showing the most consistent, strong nuclear staining in positive cases. hMSH6 was absent in two (22%) clear cell sarcomas but at least focal expression of hMLH1 and hMSH2 was observed in every case. In the melanomas, hMLH1, hMSH2 or hMSH6 was absent in four (36%), three (27%) and one (9%) case(s), respectively. Simultaneous loss of all three proteins was noted in one melanoma case (M1). The association between MSI and loss of the MMR proteins was not absolute. For example, melanoma cases M4 and M5 both demonstrated MSI-H and yet demonstrated staining for all three proteins. Conversely, clear cell sarcoma case C6 was microsatellite stable but with absent hMSH6 staining. (Figure 2k) However, a statistically significant difference between clear cell sarcoma and melanoma was observed with respect to the presence of MSI (P=0.010) and absence of hMLH1 and/or hMSH2 (P=0.014) but not hMSH6 (P=0.57).
Representative immunohistochemical results for MMR proteins in clear cell sarcoma and metastatic melanoma. MSI-stable clear cell sarcoma C2 (a) demonstrates intact hMLH1 (d), hMSH2 (g) and hMSH6 (j). MSI-stable clear cell sarcoma from patient C6 (b) demonstrates intact hMLH1 (e) and hMSH2 (h) but loss of hMSH6 immunoreactivity (k). MSI-H melanoma from patient M1 (c) demonstrates loss of immunoreactivity for all three MMR proteins with preserved staining of tumor lymphocytes (f, i, l). (a–c: H&E, 200 × ; d–f: hMLH1 immunostain, 200 × ; g–i: hMSH2 immunostain, 200 × ; j–l: hMSH6 immunostain 200 ×).
Discussion
Although clear cell sarcoma and cutaneous melanoma share similar morphologic, immunophenotypic and ultrastructural features, the histogenic relationship between the two malignancies is not completely defined. Clear cell sarcoma is characterized by a t(12;22) translocation although additional karyotypic abnormalities are sometimes observed.10, 51, 52, 53 Yet, the high degree of aneuploidy often present in melanoma is less common in clear cell sarcoma.17 In contrast to chromosomal instability, MSI is an indicator of the mutator phenotype (genetic instability) that permits the accumulation of insertion and deletion mutations in proto-oncogenes and tumor suppressor genes. Paradoxically, MSI has been observed in malignant melanoma and some karyotypically complex sarcomas, but appears to be absent in clear cell sarcoma.21, 23, 24, 44 The presence of MSI has been closely linked to defects in MMR proteins in a variety of carcinomas and some sarcomas.28, 30 In the present study, we analyzed the difference in MSI between clear cell sarcoma and large soft tissue or visceral melanoma metastases and investigated whether the differences are associated with altered expression of MMR proteins. Our study extends the previous observation that MSI is very rare in clear cell sarcoma24, 54 and goes on to show that loss of the MMR proteins hMLH1 and/or hMSH2, but not hMSH6, is correspondingly more common in metastatic melanoma than clear cell sarcoma.
The five microsatellite loci used in the present study, commonly used for analyzing colon carcinoma,47 are sufficient to detect a significant difference between clear cell sarcoma and metastatic melanoma comparable to prior studies employing a larger number of loci.24, 54 The absence of MSI in clear cell sarcoma is perhaps not surprising given the rarity of MSI in soft tissue sarcomas as a group.28 Interestingly, the subset of sarcomas displaying MSI consisted predominantly of those associated with complex karyotypes and aneuploidy but without recurrent translocations leading to unique gene-fusions (malignant fibrous histiocytoma, leiomyosarcoma).28 Although one apparent case of clear cell sarcoma with MSI has been reported, the t(12;22) status of the case was not specified.54 In keeping with the observation that loss of hMLH1 and hMSH2 is rare in most sporadic malignancies,34, 41, 55 hMLH1 and hMSH2 expression was preserved in clear cell sarcoma. However, we did note absence of hMSH6 in two clear cell sarcomas (only one of which demonstrated MSI). Loss of hMLH6 is restricted largely to germline mutations in ‘atypical’ HNPCC families that also manifest a high frequency of extracolonic malignancies, though its role in sarcomas is unknown.36, 56, 57 Germline MSH6 mutation or deletion can also produce cancer susceptibility in mice,37 but absent MSH6 expression has not been associated with sporadic malignancies.
We specifically chose to study large soft tissue or visceral melanoma metastases because such cases, in the absence of a known primary, may mimic clear cell sarcoma by routine histology, immunophenotype and ultrastructure, and pose a diagnostic challenge. It is important to recognize that the melanoma controls in the present study consisted of metastases while the clear cell sarcomas were primary tumors. Therefore, the clear cell sarcomas, despite large size and deep location, may represent an earlier stage of tumor progression than the melanomas. In the single available case of metastatic clear cell sarcoma (patient C8), we observed absence of MSI and intact MMR protein expression identical to the primary tumor (data not shown). Nevertheless, as MSI and MMR protein defects correlate with melanoma progression,23, 54, 58 it is possible that the metastatic melanoma cases are at a later stage of malignant evolution than the clear cell sarcomas. The differences in MSI and MMR protein expression may, therefore, reflect mechanistic differences in tumorigenesis or a manifestation of tumor progression.
We did not observe complete correlation between MMR protein expression and MSI. Two cases of metastatic melanoma showed intact expression of all three MMR proteins studied despite the presence of MSI. This finding may be explained by the presence of expressed, but non-functional or dominant-negative protein37 or loss of a component of the MMR system that was not studied such as hMSH3 or hPMS2.30 In contrast, one case of clear cell sarcoma demonstrated absence of hMSH6 but showed microsatellite stability at all loci studied. The findings may derive from a redundant function of hMSH6 in MMR59, 60, 61 or insufficient sensitivity of the five microsatellite loci to detect genomic instability in every case.
The difference in MSI between clear cell sarcoma and melanoma reported here and by others,24, 54 while intriguing, probably does not represent an advantage over molecular testing for t(12;22) as a diagnostic tool. However, our study shows that immunohistochemistry for hMLH1 or hMSH2 may be of diagnostic utility insofar as negative staining is compatible with melanoma but argues against clear cell sarcoma. Therefore, immunohistochemistry for these markers may serve as an adjunct to molecular techniques in difficult cases.
In summary, we have confirmed that, in contrast to metastatic melanoma, MSI is not a common feature of clear cell sarcoma. Furthermore, hMLH1 and hMSH2 expression is preserved in clear cell sarcoma. Whether these findings reflect distinct tumorigenesis pathways between these phenotypically similar malignancies, or tumor progression, remains to be determined. Immunohistochemistry for hMLH1 and hMSH2 may have utility in the diagnosis of soft tissue lesions with melanocytic differentiation.
References
Enzinger FM . Clear cell sarcoma of tendons and aponeuroses. An analysis of 21 cases. Cancer 1965;18:1163–1174.
Chung EB, Enzinger FM . Malignant melanoma of soft parts. A reassessment of clear cell sarcoma. Am J Surg Pathol 1983;7:405–413.
Granter SR, Weilbaecher KN, Quigley C, et al. Role for microphthalmia transcription factor in the diagnosis of metastatic malignant melanoma. Appl Immunohistochem Mol Morphol 2002;10:47–51.
Kindblom LG, Lodding P, Angervall L . Clear-cell sarcoma of tendons and aponeuroses. An immunohistochemical and electron microscopic analysis indicating neural crest origin. Virchows Arch A Pathol Anat Histopathol 1983;401:109–128.
Lucas DR, Nascimento AG, Sim FH . Clear cell sarcoma of soft tissues. Mayo Clinic experience with 35 cases. Am J Surg Pathol 1992;16:1197–1204.
Swanson PE, Wick MR . Clear cell sarcoma. An immunohistochemical analysis of six cases and comparison with other epithelioid neoplasms of soft tissue. Arch Pathol Lab Med 1989;113:55–60.
Montgomery EA, Meis JM, Ramos AG, et al. Clear cell sarcoma of tendons and aponeuroses. A clinicopathological study of 58 cases with analysis of prognostic factors. Int J Surg Pathol 1993;1:89–100.
Somers GR, Viero S, Nathan PC, et al. Association of the t(12;22)(q13;q12) EWS/ATF1 rearrangement with polyphenotypic round cell sarcoma of bone: a case report. Am J Surg Pathol 2005;29:1673–1679.
Barth A, Wanek LA, Morton DL . Prognostic factors in 1521 melanoma patients with distant metastases. J Am Coll Surg 1995;181:193–201.
Bridge JA, Sreekantaiah C, Neff JR, et al. Cytogenetic findings in clear cell sarcoma of tendons and aponeuroses. Malignant melanoma of soft parts. Cancer Genet Cytogenet 1991;52:101–106.
Antonescu CR, Tschernyavsky SJ, Woodruff JM, et al. Molecular diagnosis of clear cell sarcoma: detection of EWS-ATF1 and MITF-M transcripts and histopathological and ultrastructural analysis of 12 cases. J Mol Diagn 2002;4:44–52.
Patel RM, Downs-Kelly E, Weiss SW, et al. Dual-color, break-apart fluorescence in situ hybridization for EWS gene rearrangement distinguishes clear cell sarcoma of soft tissue from malignant melanoma. Mod Pathol 2005;18:1585–1590.
Speleman F, Delattre O, Peter M, et al. Malignant melanoma of the soft parts (clear-cell sarcoma): confirmation of EWS and ATF-1 gene fusion caused by a t(12;22) translocation. Mod Pathol 1997;10:496–499.
Pellin A, Monteagudo C, Lopez-Gines C, et al. New type of chimeric fusion product between the EWS and ATFI genes in clear cell sarcoma (malignant melanoma of soft parts). Genes Chromosomes Cancer 1998;23:358–360.
Garcia J, Kramer M, Mackey Z, et al. Utility of CD117 Immunoreactivity in differentiating metastatic melanoma from clear cell sarcoma. Arch Pathol Lab Med 2006;130:343–348.
Vogelstein B, Fearon ER, Kern SE, et al. Allelotype of colorectal carcinomas. Science 1989;244:207–211.
el-Naggar AK, Ordonez NG, Sara A, et al. Clear cell sarcomas and metastatic soft tissue melanomas. A flow cytometric comparison and prognostic implications. Cancer 1991;67:2173–2179.
Walker GJ, Palmer JM, Walters MK, et al. A genetic model of melanoma tumorigenesis based on allelic losses. Genes Chromosomes Cancer 1995;12:134–141.
Rao UN, Jones MW, Finkelstein SD . Genotypic analysis of primary and metastatic cutaneous melanoma. Cancer Genet Cytogenet 2003;140:37–44.
Perucho M . Microsatellite instability: the mutator that mutates the other mutator. Nat Med 1996;2:630–631.
Hussein MR . Genetic pathways to melanoma tumorigenesis. J Clin Pathol 2004;57:797–801.
Hussein MR, Sun M, Tuthill RJ, et al. Comprehensive analysis of 112 melanocytic skin lesions demonstrates microsatellite instability in melanomas and dysplastic nevi, but not in benign nevi. J Cutan Pathol 2001;28:343–350.
Palmieri G, Ascierto PA, Cossu A, et al. Assessment of genetic instability in melanocytic skin lesions through microsatellite analysis of benign naevi, dysplastic naevi, and primary melanomas and their metastases. Melanoma Res 2003;13:167–170.
Aue G, Hedges LK, Schwartz HS, et al. Clear cell sarcoma or malignant melanoma of soft parts: molecular analysis of microsatellite instability with clinical correlation. Cancer Genet Cytogenet 1998;105:24–28.
Quinn AG, Healy E, Rehman I, et al. Microsatellite instability in human non-melanoma and melanoma skin cancer. J Invest Dermatol 1995;104:309–312.
Peris K, Keller G, Chimenti S, et al. Microsatellite instability and loss of heterozygosity in melanoma. J Invest Dermatol 1995;105:625–628.
Martin SS, Hurt WG, Hedges LK, et al. Microsatellite instability in sarcomas. Ann Surg Oncol 1998;5:356–360.
Kawaguchi K, Oda Y, Takahira T, et al. Microsatellite instability and hMLH1 and hMSH2 expression analysis in soft tissue sarcomas. Oncol Rep 2005;13:241–246.
Saito T, Oda Y, Kawaguchi K, et al. Possible association between tumor-suppressor gene mutations and hMSH2/hMLH1 inactivation in alveolar soft part sarcoma. Hum Pathol 2003;34:841–849.
Kunkel TA, Erie DA . DNA mismatch repair. Annu Rev Biochem 2005;74:681–710.
Stormorken AT, Bowitz-Lothe IM, Noren T, et al. Immunohistochemistry identifies carriers of mismatch repair gene defects causing hereditary nonpolyposis colorectal cancer. J Clin Oncol 2005;23:4705–4712.
Wijnen J, de Leeuw W, Vasen H, et al. Familial endometrial cancer in female carriers of MSH6 germline mutations. Nat Genet 1999;23:142–144.
Peltomaki P, Vasen HF . Mutations predisposing to hereditary nonpolyposis colorectal cancer: database and results of a collaborative study. The International Collaborative Group on Hereditary Nonpolyposis Colorectal Cancer. Gastroenterology 1997;113:1146–1158.
Thibodeau SN, French AJ, Roche PC, et al. Altered expression of hMSH2 and hMLH1 in tumors with microsatellite instability and genetic alterations in mismatch repair genes. Cancer Res 1996;56:4836–4840.
Marcus VA, Madlensky L, Gryfe R, et al. Immunohistochemistry for hMLH1 and hMSH2: a practical test for DNA mismatch repair-deficient tumors. Am J Surg Pathol 1999;23:1248–1255.
Parc YR, Halling KC, Wang L, et al. HMSH6 alterations in patients with microsatellite instability-low colorectal cancer. Cancer Res 2000;60:2225–2231.
Yang G, Scherer SJ, Shell SS, et al. Dominant effects of an Msh6 missense mutation on DNA repair and cancer susceptibility. Cancer Cell 2004;6:139–150.
Mastrangelo D, Hadjistilianou T, Mazzotta C, et al. Microsatellite instability in three cases of embryonal rhabdomyosarcoma of the orbit. Med Pediatr Oncol 2002;39:132–133.
Rucinska M, Kozlowski L, Pepinski W, et al. High grade sarcomas are associated with microsatellite instability (chromosome 12) and loss of heterozygosity (chromosome 2). Med Sci Monit 2005;11:BR65–BR68.
Suwa K, Ohmori M, Miki H . Microsatellite alterations in various sarcomas in Japanese patients. J Orthop Sci 1999;4:223–230.
Taubert HW, Bartel F, Kappler M, et al. Reduced expression of hMSH2 protein is correlated to poor survival for soft tissue sarcoma patients. Cancer 2003;97:2273–2278.
Korabiowska M, Brinck U, Kotthaus I, et al. Analysis of the DNA content in the progression of recurrent and metastatic melanomas. Anticancer Res 2000;20:2791–2794.
Korabiowska M, Cordon-Cardo C, Jaenckel F, et al. Application of in situ hybridization probes for MLH-1 and MSH-2 in tissue microarrays of paraffin-embedded malignant melanomas: correlation with immunohistochemistry and tumor stage. Hum Pathol 2004;35:1543–1548.
Korabiowska M, Konig F, Verheggen R, et al. Altered expression and new mutations in DNA mismatch repair genes MLH1 and MSH2 in melanoma brain metastases. Anticancer Res 2004;24:981–986.
Vasen HF, Watson P, Mecklin JP, et al. New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative group on HNPCC. Gastroenterology 1999;116:1453–1456.
Boland CR, Thibodeau SN, Hamilton SR, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 1998;58:5248–5257.
Dietmaier W, Wallinger S, Bocker T, et al. Diagnostic microsatellite instability: definition and correlation with mismatch repair protein expression. Cancer Res 1997;57:4749–4756.
Dietmaier W, Riedlinger W, Kohler A, et al. Detection of microsatellite instabiltiy (MSI) and loss of heterozygosity (LOH) in colorectal tumors by fluorescence-based multiplex microsatellite PCR. Biochemica 1999;2:42–45.
Horvai AE, Li L, Xu Z, et al. Malignant mesothelioma does not demonstrate overexpression or gene amplification despite cytoplasmic immunohistochemical staining for c-Erb-B2. Arch Pathol Lab Med 2003;127:465–469.
Kang GH, Shim YH, Ro JY . Correlation of methylation of the hMLH1 promoter with lack of expression of hMLH1 in sporadic gastric carcinomas with replication error. Lab Invest 1999;79:903–909.
Fletcher JA . Translocation (12;22)(q13-14;q12) is a nonrandom aberration in soft-tissue clear-cell sarcoma. Genes Chromosomes Cancer 1992;5:184.
Bridge JA, Borek DA, Neff JR, et al. Chromosomal abnormalities in clear cell sarcoma. Implications for histogenesis. Am J Clin Pathol 1990;93:26–31.
Travis JA, Bridge JA . Significance of both numerical and structural chromosomal abnormalities in clear cell sarcoma. Cancer Genet Cytogenet 1992;64:104–106.
Richetta A, Ottini L, Falchetti M, et al. Instability at sequence repeats in melanocytic tumours. Melanoma Res 2001;11:283–289.
Misdraji J, Burgart LJ, Lauwers GY . Defective mismatch repair in the pathogenesis of low-grade appendiceal mucinous neoplasms and adenocarcinomas. Mod Pathol 2004;17:1447–1454.
Miyaki M, Konishi M, Tanaka K, et al. Germline mutation of MSH6 as the cause of hereditary nonpolyposis colorectal cancer. Nat Genet 1997;17:271–272.
Konishi M, Kikuchi-Yanoshita R, Tanaka K, et al. Molecular nature of colon tumors in hereditary nonpolyposis colon cancer, familial polyposis, and sporadic colon cancer. Gastroenterology 1996;111:307–317.
Massi D, Sardi I, Urso C, et al. Microsatellite analysis in cutaneous malignant melanoma. Melanoma Res 2002;12:577–584.
Harfe BD, Minesinger BK, Jinks-Robertson S . Discrete in vivo roles for the MutL homologs Mlh2p and Mlh3p in the removal of frameshift intermediates in budding yeast. Curr Biol 2000;10:145–148.
Marsischky GT, Filosi N, Kane MF, et al. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev 1996;10:407–420.
Kolodner RD, Marsischky GT . Eukaryotic DNA mismatch repair. Curr Opin Genet Dev 1999;9:89–96.
Acknowledgements
We thank Ms Mamie Yu and the UCSF Comprehensive Cancer Center Molecular Pathology and Immunology Cores for their technical assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
Duality of interest
None declared.
Rights and permissions
About this article
Cite this article
Garcia, J., Kramer, M., O'Donnell, R. et al. Mismatch repair protein expression and microsatellite instability: a comparison of clear cell sarcoma of soft parts and metastatic melanoma. Mod Pathol 19, 950–957 (2006). https://doi.org/10.1038/modpathol.3800611
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.3800611