Abstract
Heparanase is a mammalian endo-β-D-glucuronidase that cleaves heparan sulfate side chains at a limited number of sites. Such enzymatic activity is thought to participate in degradation and remodeling of the extracellular matrix and to facilitate cell invasion associated with tumor metastasis, angiogenesis and inflammation. Traditionally, heparanase activity was well correlated with the metastatic potential of a large number of tumor-derived cell types. More recently, heparanase upregulation has been documented in an increasing number of primary human tumors, correlating with poor postoperative survival and increased tumor vascularity. Here, we employed anti-heparanase 733 polyclonal antibody that preferentially recognizes the 50 kDa active heparanase subunit over the 65 kDa proenzyme, as well as anti-heparanase 92.4 monoclonal antibody that recognizes both the latent and the active enzyme, to follow heparanase expression, processing and localization throughout the adenoma–carcinoma transition of the colon epithelium. Normal (nondysplastic) mucosa of the large bowel near epithelial neoplasms, as well as areas of mild dysplasia in adenomas, exhibited a strong reactivity with antibody 733 that became even stronger in foci of moderate dysplasia. Interestingly, although reactivity with antibody 733 was markedly reduced in severe dysplasia and in colorectal carcinoma, response to antibody 92.4 exhibited the opposite trend and staining intensities increased in parallel with tumor stage, the highest being in carcinoma cells. Involvement of latent heparanase (detected by 92.4, but not by 733 antibody) in tumor progression was suggested by activation of the Akt/PKB signal transduction pathway upon heparanase overexpression or exogenous addition to HT29 human colon carcinoma cells. These results suggest that heparanase expression is induced during colon carcinogenesis, and that its processing, conformation and localization are tightly regulated during the course of colon adenoma–carcinoma progression.
Similar content being viewed by others
Main
Heparanase is an endoglycosidase that specifically cleaves heparan sulfate side chains of heparan sulfate proteoglycans.1, 2 Heparan sulfate proteoglycans consist of a protein core to which several heparan sulfate side chains are covalently attached. These complex macromolecules are highly abundant in the extracellular matrix and are thought to play an important structural role, contributing to extracellular matrix integrity and insolubility.3 In addition, heparan sulfate side chains can bind to a variety of biological mediators such as growth factors, cytokines and chemokines,4, 5 thus functioning as a readily available reservoir that can be liberated upon local or systemic cues. Moreover, heparan sulfate proteoglycans on the cell surface participates directly in signal-transduction cascades by potentiating the interaction between certain growth factors and their receptors.6, 7, 8, 9 Heparan sulfate-degrading activity is thus expected to affect several fundamental aspects of cell behavior under normal and clinical settings.1, 2 Traditionally, heparanase activity was implicated in cellular invasion associated with angiogenesis, inflammation and cancer metastasis.10, 11, 12, 13, 14 Recently, this notion gained further support by employing siRNA and ribozyme technologies, clearly depicting heparanase-mediated heparan sulfate cleavage and extracellular matrix remodeling as critical requisites for metastatic spread.15 Since the cloning of the heparanase gene and the availability of specific molecular probes, heparanase upregulation was documented in an increasing number of primary human tumors. Heparanase upregulation correlated with reduced postoperative survival of pancreatic,16 bladder,17 gastric,18 cervical19 and colorectal20 cancer patients. Similarly, heparanase upregulation correlated with increased lymph node and distant metastases,17, 18, 21, 22, 23, 24, 25 providing a strong clinical support for the prometastatic feature of heparanase.
The heparanase cDNA encodes for a protein of 543 amino acids that undergoes proteolytic processing at two potential cleavage sites, Glu109–Ser110 and Gln157–Lys158, yielding an 8 kDa polypeptide at the N-terminus and a 50 kDa polypeptide at the C-terminus that heterodimerize to form an active heparanase enzyme.26, 27, 28 Recently, we have generated a polyclonal antibody (733) that preferentially recognizes the 50 kDa heparanase subunit vs the 65 kDa latent enzyme, and used this antibody to study the trafficking routes, processing and localization of heparanase in vitro.29 Furthermore, this antibody is suitable for staining of paraffin sections.29 Thus, immunostaining of archival material can be correlated with heparanase enzymatic activity. We hypothesized that heparanase expression and activity are induced during colon carcinogenesis, and employed antibody 733, and monoclonal antibody 92.4 that recognizes both the latent and active forms of the enzyme, to follow heparanase expression pattern, processing and activation during the adenoma–carcinoma sequence of the colon. We provide evidence that normal-looking colon epithelium away (0.5–1 cm) from the tumor is negative for both antibodies. Normal (nondysplastic) mucosa of the large bowel near epithelial neoplasms, as well as areas of mild dysplasia in adenomas, exhibited a strong reactivity with antibody 733 that became even stronger in foci of moderate dysplasia. Interestingly, although reactivity with antibody 733 was markedly reduced in severe dysplasia and in colorectal carcinoma, response to antibody 92.4 exhibited the opposite trend and staining intensities increased in parallel with tumor stage, the highest being in carcinoma cells. These results suggest that heparanase expression is induced during colon carcinogenesis, and also imply that processing and localization of the enzyme are tightly regulated during the course of colon adenoma–carcinoma progression.
Materials and methods
Specimen Selection Strategy
This study was conducted on specimens of colorectal villous adenomas (25 patients) and colorectal carcinomas (38 patients), who had no history consistent with hereditary nonpolyposis colorectal cancers, familial adenomatous polyposis or inflammatory bowel disease, undergoing surgical resection at the Hadassah–Hebrew University Hospital. We assessed the expression of heparanase in cells from individual microscopic fields, representing the gradient of dysplastic changes characteristic to the adenoma–carcinoma sequence route, rather than compared the expression of heparanase in the different colorectal epithelial tumors. Selection of the specimens was conducted accordingly. As foci of high-grade dysplasia, carcinoma in situ and invasive cancer are most prevalent in villous colorectal adenomas, the adenoma specimens selected were of this type. Of the 25 villous adenoma cases, seven presented with foci of invasive carcinoma and 16 were diagnosed as high-grade adenomas. Specimens from the other two adenoma types (tubular and tubulovillous) were not included, thus ensuring the highest possible yield of microscopic fields from the various histological entities relevant to the study.
For each case, tissue samples of the tumor lesion and of normal mucosa distant from the tumor were included in the study. Three different areas of the specimen were examined in each case. They consisted of a segment of the tumor from the periphery, and the normal-looking mucosa at its border, as well as samples from deeper areas of the tumor, including the deepest growing edge. Staining was performed on paraffin-embedded sections of all samples. In addition, 25% of the specimens were freshly obtained and processed as frozen sections. Microscopic fields (>100 cells/field) selected for evaluation from all neoplastic samples were categorized by three expert pathologists (VD, BM and YS) according to the criteria of the Vienna Classification. The noninvasive low-grade neoplasia category (Category 3) was further subdivided into mild and moderate dysplasia subgroups, based upon accepted histological criteria. Invasive carcinomas (Category 5) were further divided into two subgroups according to depth of invasion into the surrounding stroma. The first subgroup consisted of microscopic fields from the superficial layers of the invasive carcinoma and the second subgroup consisted of fields from the growing edge, invading deep into the neighboring tissues. The corresponding normal mucosas were included as a control for the neoplasia, and were further subdivided into three subgroups according to the distance from tumor (Table 1).
Antibodies and Immunohistochemistry
Antibody 733 was raised in rabbits against a 15-amino-acid peptide (KKFKNSTYSRSSVDC) that maps at the C-terminus of the 50 kDa heparanase subunit. This antibody has previously been shown to recognize preferentially the 50 kDa subunit over the latent 65 kDa heparanase protein as revealed by immunoblot analysis, and to stain positively heparanase-expressing cytotrophoblasts of the placenta by immunohistochemistry.29 Anti-heparanase monoclonal antibody 92.4 has previously been characterized and shown to stain positively paraffin-embedded colon carcinoma specimens.30, 31 Antibody 1453 was raised in rabbits against the full-length 65 kDa latent heparanase protein.26 Immunohistochemistry was performed essentially as described.31, 32 Briefly, 5 μm sections were deparaffinized and rehydrated, followed by antigen retrieval (boiling in 10 mM citrate buffer, pH 6, for 10 min in a microwave oven). Sections were then incubated with 3% H2O2 in absolute methanol for 30 min to inactivate endogenous peroxidase activity, washed with PBS and incubated with 10% goat serum to block nonspecific binding, followed by overnight incubation with the respective primary antibody (pAb 733 diluted 1:100, mAb 92.4 diluted 1:5 in blocking solution) at 4°C. Sections were then incubated (30 min, RT) with biotin-conjugated anti-rabbit IgG or anti-mouse IgM secondary antibodies, respectively, and were then incubated (30 min, RT) with avidin–peroxidase reagent, according to the manufacturer's (Zymed Laboratories, San Francisco, CA, USA) instructions. Finally, color was developed by the AEC chromogen (Zymed), and sections were counterstained with hematoxylin and mounted. PBS containing 0.1% Triton X-100 was used to extensively wash the slides after each step of the staining procedure. Staining of 5 μm acetone-fixed frozen sections was similarly performed.
Scoring Methodology
All immunostained slides were examined by three expert pathologists (VD, BM and YS), using a multiheaded microscope. Microscopic fields for evaluation were selected in agreement by all three pathologists, whereas the histological grading and staining intensity scoring were performed independently in a blind manner. Any interobserver variability was resolved by consensus at the multiheaded microscope. Immunostaining scoring and histological categorization was documented for all microscopic fields examined. Irrelevant to morphological definition of the original specimens, the randomly chosen fields from all selected cases were reassigned to new groups, each of which represented one of the histological grades characteristic of the various stages of adenoma–carcinoma sequence and the overall staining score of each group was evaluated. Staining was scored on a scale of 0–4 according to the number of positive tumor cells, as follows: (0) 0%; (1) <10%; (2) 10–50%; (3) 51–80% and (4) >80%. Intensity of staining was also rated on a scale of 0–3, with different intensity categories implemented for each of the two antibodies examined, owing to their marked differences in staining patterns. Intensity categories for antibody 733 (supranuclear aggregates) were: (0) no staining; (1) weak staining, observed at ≥ × 400 magnification; (2) moderate staining, observed at ≥ × 100 magnification and (3) strong staining, observed at × 50 magnification. Intensity categories for antibody 92.4 (dispersed cytoplasmic and membranous) were: (0) no staining; (1) weak staining, barely observed over nonspecific background staining; (2) moderate staining observed as coarse aggregates, partially obscuring cytoplasm and counterstained chromatin and (3) strong staining, observed as coarse membranous bands obscuring the cell boundaries. The values of the two parameters (% positive cells, staining intensity) for all microscopic fields examined were multiplied, resulting in scores ranging from 0 to 12 (Remmele score (RS), 32), which were further subdivided into six intensity categories as follows: −, RS=0; ±, RS=1; +, RS=2, ++, RS=3–4; +++, RS=6–9 and ++++, RS=12.
Cells, Cell Culture and Transfection
The HT29 human colon carcinoma cell line was purchased from the American Tissue Culture Collection (ATCC). Cells were grown in Dulbecco's modified Eagle's medium supplemented with glutamine, pyruvate, antibiotics and 10% fetal calf serum in a humidified atmosphere containing 8% CO2, at 37°C. For stable transfection, cells were transfected with the pSecTag 2 vector containing the full-length heparanase cDNA, kindly provided by Dr Hua-Quan Miao (ImClone Systems Inc., New York, NY, USA),28 using the jetPEI reagent according to the manufacturer's (Poly transfection, Illkrich Cedex, France) instructions, selected with Zeocin (Invitrogen, Carlsbad, CA, USA) for 3 weeks, expanded and pooled.
Cell Lysates and Protein Blotting
Cell cultures were incubated for 20 h under serum-free conditions, pretreated with 1 mM orthovanadate for 10 min at 37°C, washed twice with ice-cold PBS containing 1 mM orthovanadate and scraped into lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, 1 mM orthovanadate, 1 mM PMSF) containing a cocktail of proteinase inhibitors (Roche, Mannheim, Germany). Total cellular protein concentration was determined by the BCA assay, according to the manufacturer's (Pierce, Rockford, IL, USA) instructions. Cellular proteins (30 μg) were fractionated on SDS-PAGE and immunoblotting was performed essentially as described.32, 33, 34 The following primary antibodies were employed: anti-heparanase (1453), anti-phosphorylated Akt (Cell Signaling, Beverly, MA, USA), anti-Akt (Santa Cruz, CA, USA) and anti-actin (Sigma, Saint Louis, MO, USA).
Tumorigenicity
Cells from exponential cultures of control (Vo) or heparanase transfected HT29 cells were detached with trypsin, washed with PBS and brought to a concentration of 5 × 107 cells/ml. Cell suspension (5 × 106/0.1 ml) was inoculated subcutaneously at the right flank of 5-week-old female SCID mice (n=5). Xenograft size was determined twice a week by externally measuring tumors in two dimensions using a caliper. Tumor volume (V) was determined by the equation V=L × W2 × 0.5, where L is the length and W the width of the xenograft. At the end of the experiment, mice were killed, and xenografts were resected, weighed, fixed in formalin and 5 μm sections were stained with hematoxylin–eosin.
Heparanase Activity Assay
Preparation of extracellular matrix-coated 35 mm dishes and determination of heparanase activity were performed as described in detail elsewhere.30, 34 To evaluate heparanase enzymatic activity, control Vo and heparanase-transfected cells (1 × 106) were lysed by three freeze/thaw cycles and the resulting cell extracts were incubated (18 h, 37°C, pH 6.0) with 35S-labeled extracellular matrix. Alternatively, cells were left intact and plated directly into dishes coated with 35S-labeled extracellular matrix. The incubation medium (1 ml) containing sulfate-labeled degradation fragments was subjected to gel filtration on a Sepharose CL-6B column. Fractions (0.2 ml) were eluted with PBS and their radioactivity counted in a λ-scintillation counter. Degradation fragments of heparan sulfate side chains generated by heparanase are eluted at 0.5<Kav<0.8 (peak II, fractions 15–30). Nearly intact heparan sulfate proteoglycans are eluted just after the Vo (Kav<0.2, peak I, fractions 3–15).
Statistical Analysis
The Statistical toolbox, version 5, of the MATLAB 7.1.0.246 (R14) Service pack 3 program, was used for statistical analyses. Association between heparanase expression and the histological grading was evaluated by rank correlation using Mann–Whitney U-test and Wilcoxon two-sided rank-sum test. P-values<0.05 were considered statistically significant.
Results
In all specimens examined, normal mucosa far away (0.5–1 cm) from the tumor exhibited no reactivity with the anti-heparanase antibodies employed in this study (Figures 1a, 2a, Table 1). Normal-looking mucosa closer (0.2–0.5 cm) to the carcinoma, as well as that bordering the carcinoma (transitional mucosa), showed a detectable and consistent reactivity with the anti-heparanase antibodies (Figures 1b, 2b, Table 1), and exhibited a stronger staining intensity, approaching the cancerous lesion (Table 1; P≪0.05).
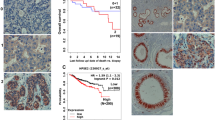
Heparanase expression in epithelial cells of colon mucosa in the various stages of adenoma–carcinoma sequence, as depicted by anti-heparanase antibody 92.4. (a) Normal mucosa, distant from the tumor, devoid of heparanase staining. Original magnification × 100. (b) Normal mucosa (arrowheads) adjacent to carcinoma (arrows), exhibiting a detectable staining for heparanase. Original magnification × 50. (c) Part of an adenoma. Colon tissue with mild dysplasia exhibits enhanced cytoplasmic staining for heparanase. Original magnification × 400. (d) Colon carcinoma exhibiting strong heparanase staining. Note that heparanase is being diffusely localized in the cytoplasm of the carcinoma cells, and also assume membrane-like condensation, in the circumference of the tumor fragments. Original magnification × 200. (e) Clusters of invading carcinoma cells at the growing edge (arrowheads) showing increased staining intensity, as compared with noninvading tumor cells. Original magnification × 100. (f) Small clusters (arrows) and single cells (arrowheads) of invading carcinoma, showing marked concentration of heparanase in a membrane-like localization. Original magnification × 400.
Heparanase expression in epithelial cells of colon mucosa in the various stages of adenoma–carcinoma sequence, as depicted by anti-heparanase antibody 733. (a) Normal mucosa, distant from the tumor, devoid of heparanase staining (original magnification × 100). (b) Normal mucosa (arrowheads) adjacent to carcinoma, exhibiting supranuclear staining for heparanase (inset 1). Original magnification × 50. (c) Adenoma exhibiting strong, widespread supranuclear staining (inset). Original magnification × 100. (d) A part of an adenoma. Heparanase staining is much more intense in areas with moderate dysplasia (arrowheads), as compared to areas with mild or no dysplasia (arrows). Original magnification × 200. (e) Same as in (d). Heparanase staining intensity in area with severe dysplasia (arrows) is weaker, as compared to an area with moderate dysplasia (arrowheads). Original magnification × 400). (f) Invading carcinoma exhibiting very weak staining in a few isolated cells. Original magnification × 200.
Expression and Localization of Heparanase, as Evident by Antibody 92.4
Reactivity with antibody 92.4, recognizing both the latent and active heparanase forms, was evident in normal mucosa adjacent to the tumor. Similar staining intensities were observed in microscopic fields of mild and moderate dysplasia (Figure 1c). Staining intensity significantly increased in severe dysplasia (Table 1; P≪0.05) and a further increase in reactivity was observed in colorectal carcinoma, regardless of its differentiation status and whether in situ or invasive (Figure 1d). By far, the strongest reactivity with antibody 92.4 was observed in the deep growing edge of the carcinoma (Figure 1e, arrowheads, Table 1; P≪0.05 compared to carcinoma at a more superficial level), in agreement with the proinvasive function of the heparanase enzyme.2, 15, 30, 31, 35 In cell clusters, heparanase was noted to assume both diffuse cytoplasmic and membrane-like localization (Figure 1f, arrows), whereas invading single carcinoma cells exhibited mostly membrane-like localization of heparanase (Figure 1f, arrowheads).
Expression and Localization of Heparanase, as Evident by Antibody 733
Similar to the staining with antibody 92.4, reactivity with antibody 733, which preferentially recognizes the 50 kDa active heparanase protein, was noted in normal-looking mucosa relatively close (0.2–0.5 cm) to the tumor (Figure 2b, arrowheads and inset 1). A marked increase in staining intensity was noted in areas with moderate dysplasia (Figures 2d, e, arrowheads, P≪0.05; Table 1). Surprisingly, severe dysplasia (Figure 2e, arrows) and, even more so, carcinoma of the colon (Figures 2b (inset 2) and 2f) exhibited a significant reduction in reactivity with antibody 733 (Table 1; P≪0.05). In contrast with the diffuse cytoplasmic and membrane localization noted with antibody 92.4, staining with antibody 733 mainly revealed localization of heparanase to the supranuclear aspect of colon epithelia in a pattern that somewhat resembled vesicle-like structures (Figures 2b (inset 1) and 2c (inset)). These vesicles have been previously characterized as endocytic endosomes and lysosomes in cells maintained in vitro, as well as in tumor xenografts.29, 36, 37 Thus, the two antibodies employed in this study stained and distinguished between different heparanase pools at the course of colon carcinoma progression, suggesting that heparanase expression, processing and localization is dynamic and tightly regulated.
Heparanase Overexpression Facilitates HT29 Xenograft Growth
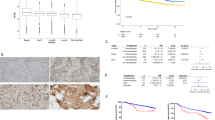
In order to study the contribution of heparanase to colon tumor progression, heparanase was stably transfected into HT29 human colon carcinoma cells. Transfected HT29 cells exhibited high levels of heparanase activity in cell lysates (Figure 3a), in accordance with high expression levels of heparanase revealed by immunoblot analysis with anti-heparanase antibodies (Figure 3a, inset). Moreover, high levels of heparanase activity were detected upon plating intact cells on dishes coated with 35S-labeled extracellular matrix (Figure 3b), suggesting that heparanase is efficiently secreted by these cells. Indeed, heparanase was readily detected in the culture medium of heparanase-transfected HT29 cells (Figure 3b, inset). Previously, we have noted elevated levels of Akt/PKB phosphorylation upon heparanase overexpression or exogenous addition,32, 33 suggesting that heparanase is involved in signaling cascades. Subjecting cell lysates to immunoblotting with antibody directed against the phosphorylated state of Akt/PKB clearly revealed a marked increase of Akt phosphorylation in heparanase-overexpressing cells (Figure 3c, Hepa, left panel) compared with control Vo cells (Figure 3c, Vo, left panel). Similarly, exogenous addition of recombinant heparanase (1 μg/ml) to HT29 cell cultures stimulated Akt phosphorylation (Figure 3c, right panel) comparable in magnitude to heparanase overexpression (Figure 3c, left panel), further supporting the notion that heparanase is capable of initiating signal transduction. Next, control Vo and heparanase overexpressing HT29 cells were implanted subcutaneously in immune-deficient SCID mice and tumor growth was monitored during 24 days. Heparanase-overexpressing cells produced xenografts threefold larger in volume (Figure 3d) and 2.5-fold bigger in weight (Figure 3e) compared with xenografts produced by control Vo cells. These results indicate that heparanase enhances colon xenograft growth, possibly through the activation of the Akt/PKB pathway that is strongly implicated in cell survival.
Heparanase overexpression induces Akt/PKB phosphorylation and facilitates xenograft growth. (a, b) Heparanase activity. Heparanase cDNA was stably transfected into HT29 cells. Control Vo (♦) and heparanase-transfected (▴) cells (1 × 106) were plated intact on 35S-labeled extracellular matrix (b) or subjected to three cycles of freeze/thaw before incubation (18 h, 37°C, pH 6.0) with the labeled extracellular matrix. (a). Sulfate-labeled degradation fragments released into the incubation medium were subjected to gel filtration on Sepharose 6B, as described in ‘Materials and methods’. (Insets) Heparanase expression and secretion. Cell lysates (a) and conditioned medium (b) of control Vo and heparanase-transfected (Hepa) cells were immunoblotted with anti-heparanase 1453 antibodies to reveal heparanase expression (a) and secretion into the culture medium (b). (c) Akt/PKB phosphorylation. Lysates of control (Vo) and heparanase-transfected (Hepa) HT29 cells were immunoblotted with anti-phospho (left, upper panel) and total (left, lower panel) Akt/PKB antibodies. HT29 cells were left untreated (0) or incubated with recombinant heparanase (1 μg/ml) for the time indicated (min) and cell lysates were immunoblotted with antibody directed against phosphorylated (right, upper panel) and total (right, lower panel) Akt/PKB. (d, e) Xenograft development. (d) Tumor volume. Control Vo and heparanase-overexpressing HT29 cells (5 × 106) were implanted subcutaneously at the flank of SCID mice (n=5). Xenograft size was measured twice a week with a caliper and tumor volume was calculated. Tumor weight (g) measured at the end of the experiment on day 62 is shown in (e). A 2.5-fold increase in tumor weight is noted in tumors produced by heparanase-overexpressing cells.
Discussion
In addition to its intimate involvement in the egress of cells from the blood stream, heparanase activity may release a multitude of heparan sulfate-bound, extracellular matrix-resident growth factors, cytokines, chemokines and enzymes that might profoundly affect cell and tissue function. Thus, heparanase expression, activity and bioavailability should be kept tightly regulated. Mechanisms that dictate heparanase regulation are only poorly understood, but expected to operate at several distinct levels. Heparanase overexpression in multiple human tumors suggests a transcriptional regulation. Heparanase gene expression has been shown to involve promoter methylation,38 eukaryotic initiation factor 4E,39 and the ETS40 and Egr141 transcription factors. Regulation at the post-translational level, namely heparanase processing, cellular localization and secretion, has also been implicated as major regulatory mechanisms.26, 27, 28, 42 Heparanase upregulation has been noted in several primary human tumors, including colon carcinoma by employing in situ hybridization and immunostaining.31 Moreover, heparanase upregulation in colon carcinoma correlated with increased lymph vessel and vascular infiltration and, most importantly, with reduced postoperative survival rate,43 suggesting that heparanase expression levels may be considered as an important diagnostic parameter. The antibodies so far employed for heparanase detection do not distinguish, however, between the latent 65 kDa and active 50 kDa forms of the enzyme. Here, we utilized polyclonal antibody 733 and monoclonal antibody 92.4 that preferentially recognize the active or both the latent and active forms of heparanase, respectively, to follow expression levels, processing and localization of heparanase during the adenoma–carcinoma sequence in the colon. Reactivity with the 92.4 monoclonal antibody increased substantially during the transition from mild dysplasia to carcinoma, indicating induced heparanase expression during colon carcinogenesis, and in agreement with our previous observations.31 Surprisingly, reactivity with antibody 733 exhibited the opposite trend. Most remarkably, antibody 733 hardly detected heparanase in colorectal carcinoma, but efficiently detected heparanase upregulation in the progression from normal epithelia to moderate dysplasia. We have recently utilized the human glioma U87 cell line to study the contribution of heparanase to xenograft tumor progression.32 Interestingly, although low levels of heparanase significantly enhanced xenograft growth, high heparanase levels exhibited the opposite effect and inhibited xenograft development.32 This result suggests that in certain tumors, attenuated heparanase expression levels are associated with increased tumor growth. Indeed, heparanase was readily detected in hyperplastic prostate tissue, but rarely detected in prostate carcinoma graded as Gleason ≥7,44 further supporting such a possibility and depicting the complexity of heparanase regulation. The situation in colorectal carcinoma may provide another level of complexity. Clearly, heparanase expression was not attenuated in colorectal carcinoma, but rather heparanase processing, as evident by the decreased reactivity with antibody 733, shown to recognize preferentially the active 50 kDa subunit of the enzyme. Recently, we have demonstrated that exogenous addition of the latent 65 kDa heparanase stimulates Akt-dependent endothelial cell invasion and migration,33 presumably independent of heparanase enzymatic activity. Nonenzymatic activities of heparanase also include enhanced adhesion of tumor-derived cells32, 45 and primary T cells,46 mediated by β1-integrin activation and correlated with Akt, Pyk2 and ERK activation.32, 46 More recently, we have demonstrated that heparanase is intimately involved in the regulation of vascular endothelial growth factor expression, thus contributing to tumor angiogenesis.47 Vascular endothelial growth factor induction required secretion of heparanase but not enzymatic activity, and was found to be mediated by Src activation.47 It is thus possible that heparanase activity, evident by reactivity with antibody 733, is required for the transition from normal colon epithelium to low-grade adenoma, but that further progression to carcinoma is associated with a decrease in heparanase activity. Heparanase may still contribute to tumor progression at this stage through its signaling capabilities, and its localization evident by antibody 92.4 staining may suggest that heparanase-mediated signal transduction is initiated at the cell membrane, possibly interacting with heparanase receptor. Indeed, elevated levels of Akt/PKB phosphorylation were observed upon heparanase overexpression or exogenous addition to HT29 colon carcinoma cells, in correlation with an enhanced xenograft growth. These results suggest that heparanase is capable of initiating signal transduction, supporting migration, proliferation and survival of colon carcinoma cells. Studies utilizing antibodies 733 and 92.4 to examine heparanase expression, processing and cellular localization in other human tumors are currently in progress.
References
Vlodavsky I, Friedmann Y . Molecular properties and involvement of heparanase in cancer metastasis and angiogenesis. J Clin Invest 2001;108:341–347.
Parish CR, Freeman C, Hulett MD . Heparanase: a key enzyme involved in cell invasion. Biochim Biophys Acta 2001;1471:M99–M108.
Kjellen L, Lindahl U . Proteoglycans: structures and interactions. Annu Rev Biochem 1991;60:443–475.
Capila I, Linhardt RJ . Heparin–protein interactions. Angew Chem Int Ed Engl 2002;41:391–412.
Munoz EM, Linhardt RJ . Heparin-binding domains in vascular biology. Arterioscler Thromb Vasc Biol 2004;24:1549–1557.
Eswarakumar VP, Lax I, Schlessinger J . Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev 2005;16:139–149.
Raab G, Klagsbrun M . Heparin-binding EGF-like growth factor. Biochim Biophys Acta 1997;1333:F179–F199.
Tanaka Y, Kimata K, Adams DH, et al. Modulation of cytokine function by heparan sulfate proteoglycans: sophisticated models for the regulation of cellular responses to cytokines. Proc Assoc Am Physicians 1998;110:118–125.
Vlodavsky I, Miao HQ, Atzmon R, et al. Control of cell proliferation by heparan sulfate and heparin-binding growth factors. Thromb Haemost 1995;74:534–540.
Vlodavsky I, Fuks Z, Bar-Ner M, et al. Lymphoma cells mediated degradation of sulfated proteoglycans in the subendothelial extracellular matrix: relation to tumor cell metastasis. Cancer Res 1983;43:2704–2711.
Nakajima M, Irimura T, DiFerrante D, et al. Heparan sulfate degradation: relation to tumor invasion and metastatic properties of Mouse B 16 Melanoma sublines. Science (Washington, DC) 1983;220:611–613.
Nakajima M, Irimura T, Di Ferrante N, et al. Metastatic melanoma cell heparanase. Characterization of heparan sulfate degradation fragments produced by B16 melanoma endoglucuronidase. J Biol Chem 1984;259:2283–2290.
Naparstek Y, Cohen IR, Fuks Z, et al. Activated T lymphocytes produce a matrix-degrading heparan sulphate endoglycosidase. Nature 1984;310:241–244.
Matzner Y, Bar-Ner M, Yahalom J, et al. Degradation of heparan sulfate in the subendothelial extracellular matrix by a readily released heparanase from human neutrophils. Possible role in invasion through basement membranes. J Clin Invest 1985;76:1306–1313.
Edovitsky E, Elkin M, Zcharia E, et al. Heparanase gene silencing, tumor invasiveness, angiogenesis, and metastasis. J Natl Cancer Inst 2004;96:1219–1230.
Koliopanos A, Friess H, Kleeff J, et al. Heparanase expression in primary and metastatic pancreatic cancer. Cancer Res 2001;61:4655–4659.
Gohji K, Hirano H, Okamoto M, et al. Expression of three extracellular matrix degradative enzymes in bladder cancer. Int J Cancer 2001;95:295–301.
Takaoka M, Naomoto Y, Ohkawa T, et al. Heparanase expression correlates with invasion and poor prognosis in gastric cancers. Lab Invest 2003;83:613–622.
Shinyo Y, Kodama J, Hongo A, et al. Heparanase expression is an independent prognostic factor in patients with invasive cervical cancer. Ann Oncol 2003;14:1505–1510.
Sato T, Yamaguchi A, Goi T, et al. Heparanase expression in human colorectal cancer and its relationship to tumor angiogenesis, hematogenous metastasis, and prognosis. J Surg Oncol 2004;87:174–181.
Watanabe M, Aoki Y, Kase H, et al. Heparanase expression and angiogenesis in endometrial cancer. Gynecol Obstet Invest 2003;56:77–82.
Chen JQ, Zhan WH, He YL, et al. Expression of heparanase gene, CD44v6, MMP-7 and nm23 protein and their relationship with the invasion and metastasis of gastric carcinomas. World J Gastroenterol 2004;10:776–782.
Maxhimer JB, Quiros RM, Stewart R, et al. Heparanase-1 expression is associated with the metastatic potential of breast cancer. Surgery 2002;132:326–333.
Rohloff J, Zinke J, Schoppmeyer K, et al. Heparanase expression is a prognostic indicator for postoperative survival in pancreatic adenocarcinoma. Br J Cancer 2002;86:1270–1275.
Tang W, Nakamura Y, Tsujimoto M, et al. Heparanase: a key enzyme in invasion and metastasis of gastric carcinoma. Mod Pathol 2002;15:593–598.
Fairbanks MB, Mildner AM, Leone JW, et al. Processing of the human heparanase precursor and evidence that the active enzyme is a heterodimer. J Biol Chem 1999;274:29587–29590.
McKenzie E, Young K, Hircock M, et al. Biochemical characterisation of the active heterodimer form of Human Heparanase (Hpa1) protein expressed in insect cells. Biochem J 2003;373:423–435.
Levy-Adam F, Miao HQ, Heinrikson RL, et al. Heterodimer formation is essential for heparanase enzymatic activity. Biochem Biophys Res Commun 2003;308:885–891.
Zetser A, Levy-Adam F, Kaplan V, et al. Processing and activation of latent heparanase occurs in lysosomes. J Cell Sci 2004;117:2249–2258.
Vlodavsky I, Friedmann Y, Elkin M, et al. Mammalian heparanase: gene cloning, expression and function in tumor progression and metastasis. Nat Med 1999;5:793–802.
Friedmann Y, Vlodavsky I, Aingorn H, et al. Expression of heparanase in normal, dysplastic, and neoplastic human colonic mucosa and stroma. Evidence for its role in colonic tumorigenesis. Am J Pathol 2000;157:1167–1175.
Zetser A, Bashenko Y, Miao H-Q, et al. Heparanase affects adhesive and tumorigenic potential of human glioma cells. Cancer Res 2003;63:7733–7741.
Gingis-Velitski S, Zetser A, Flugelman MY, et al. Heparanase induces endothelial cell migration via protein kinase B/Akt activation. J Biol Chem 2004;279:23536–23541.
Levy-Adam F, Abboud-Jarrous G, Guerrini M, et al. Identification and characterization of heparin/heparan sulfate binding domains of the endoglycosidase heparanase. J Biol Chem 2005;280:20457–20466.
Dempsey LA, Plummer TB, Coombes SL, et al. Heparanase expression in invasive trophoblasts and acute vascular damage. Glycobiology 2000;10:467–475.
Nadav L, Eldor A, Yacoby-Zeevi O, et al. Activation, processing and trafficking of extracellular heparanase by primary human fibroblasts. J Cell Sci 2002;115:2179–2187.
Goldshmidt O, Nadav L, Aingorn H, et al. Human heparanase is localized within lysosomes in a stable form. Exp Cell Res 2002;281:50–62.
Shteper PJ, Zcharia E, Ashhab Y, et al. Role of promoter methylation in regulation of the mammalian heparanase gene. Oncogene 2003;22:7737–7749.
Yang YJ, Zhang YL, Li X, et al. Contribution of eIF-4E inhibition to the expression and activity of heparanase in human colon adenocarcinoma cell line: LS-174T. World J Gastroenterol 2003;9:1707–1712.
Lu WC, Liu YN, Kang BB, et al. Trans-activation of heparanase promoter by ETS transcription factors. Oncogene 2003;22:919–923.
de Mestre AM, Khachigian LM, Santiago FS, et al. Regulation of inducible heparanase gene transcription in activated T cells by early growth response 1. J Biol Chem 2003;278:50377–50385.
Goldshmidt O, Zcharia E, Abramovitch R, et al. Cell surface expression and secretion of heparanase markedly promote tumor angiogenesis and metastasis. Proc Natl Acad Sci USA 2002;99:10031–10036.
Nobuhisa T, Naomoto Y, Ohkawa T, et al. Heparanase expression correlates with malignant potential in human colon cancer. J Cancer Res Clin Oncol 2005;131:229–237.
Stadlmann S, Moser PL, Pollheimer J, et al. Heparanase-1 gene expression in normal, hyperplastic and neoplastic prostatic tissue. Eur J Cancer 2003;39:2229–2233.
Goldshmidt O, Zcharia E, Cohen M, et al. Heparanase mediates cell adhesion independent of its enzymatic activity. FASEB J 2003;17:1015–1025.
Sotnikov I, Hershkoviz R, Grabovsky V, et al. Enzymatically quiescent heparanase augments T cell interactions with VCAM-1 and extracellular matrix components under versatile dynamic contexts. J Immunol 2004;172:5185–5193.
Zetser A, Bashenko Y, Edovitsky E, et al. Heparanase induces VEGF expression: Correlation with p38 phosphorylation levels and Src activation. Cancer Res 2006;66:1455–1463.
Acknowledgements
We would like to thank InSight Biopharmaceutical Ltd (Rabin Science Park, Rehovot, Israel) for providing the anti-heparanase 92.4 monoclonal antibody. This work was supported by Grant 532/02 from the Israel Science Foundation; by United States Public Health Service Grant RO1 CA106456 from NCI, National Institutes of Health; and by a grant in memory of Herbert and Frances Brody from Harvey M Krueger, as trustee of a charitable trust.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Doviner, V., Maly, B., Kaplan, V. et al. Spatial and temporal heparanase expression in colon mucosa throughout the adenoma-carcinoma sequence. Mod Pathol 19, 878–888 (2006). https://doi.org/10.1038/modpathol.3800603
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.3800603