Abstract
Cytologic evaluation of ductal lavage or random periareolar fine-needle aspiration (FNA) specimens has been proposed to improve risk stratification of women at high risk for breast cancer. However, cytologic assessment of morphologic changes is subjective. To assess the utility of fluorescence in situ hybridization (FISH) in the categorization of breast lesions, we prospectively evaluated 32 random periareolar FNA specimens from 27 women at high risk for breast cancer. Cytologic specimens were prepared using the thin preparation technique, and diagnoses were made on the basis of previously published criteria. Specimens were also evaluated by FISH for chromosomes 1, 8, 11, and 17. Monosomy was defined as the loss of one signal or both signals in >20% of cells, and polysomy was defined as the presence of ≥3 signals in >6% of cells. Cytologic smears from seven invasive ductal carcinomas and nine benign breast specimens from women at low risk for breast cancer were included for comparison. In the high-risk group, cytologic findings were nonproliferative epithelium (NPE) in 16 cases and hyperplasia in 16 cases. Chromosomal aberrations were detected in 11 (69%) of 16 NPE cases, 14 (89%) of 16 hyperplasia cases, seven (100%) of seven carcinoma cases, and none of the low-risk cases. High-risk cases had significantly more monosomy of chromosomes 1, 11, and 17 and polysomy of chromosome 8 compared to low-risk cases and significantly less polysomy of chromosomes 1, 8, 11, and 17 compared to patients with cancer. There were no significant differences in monosomy or polysomy of individual chromosomes or a combination of chromosomes between the NPE and hyperplasia groups. This study shows that aberrations of chromosome number are common in high-risk women irrespective of cytologic findings. Studies evaluating the association between specific patterns of chromosomal polysomy and progression to malignancy may be warranted.
Similar content being viewed by others
Main
Cytologic evaluation of ductal lavage and periareolar fine-needle aspiration (FNA) specimens has recently been advocated as a new modality for improving the risk stratification of women at high risk for breast cancer.1, 2 In the few studies that have examined the breast cancer risk associated with cytologic atypia, a finding of cytologic atypia was associated with a breast cancer risk similar to that reported in women with a finding of atypical hyperplasia on examination of histologic sections.2, 3
Although the cytologic criteria used to categorize intraductal epithelial changes are well defined, they have not been uniformly applied, and they have not been proven to be reproducible.4, 5 In studies that have compared cytologic findings of atypia with histologic findings from corresponding tissue, the correlation was poor and the rate of false-positive results high.1, 5 Consequently, ancillary studies to better assess cellular changes are being sought.6
Breast cancer progression is characterized by the accumulation of numeric changes on many chromosomes, particularly early or frequent increases in the copy numbers of chromosomes 1, 8, 11, and 17.7, 8, 9, 10, 11, 12, 13, 14, 15, 16 Fluorescence in situ hybridization (FISH) evaluation of chromosomal aberrations in ductal lavage specimens may be more sensitive and specific than conventional cytologic evaluation for categorization of breast lesions.17, 18, 19 To determine the utility of FISH in categorizing breast epithelial changes, we used this technique to evaluate numeric changes in chromosomes 1, 8, 11, and 17 in breast FNA specimens from 27 women at high risk for breast cancer. For comparison, we used cytologic smears from seven invasive ductal breast carcinomas and nine specimens of nonproliferative epithelium (NPE) from women at low risk for breast cancer.
Materials and methods
Fine-Needle Aspiration
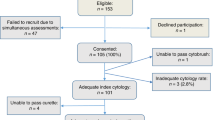
The patients from whom FNA specimens were obtained had been enrolled in an institutional review board-approved prospective protocol to determine the feasibility of cytologic evaluation and evaluation of biomarkers in ductal lavage and periareolar FNA specimens for stratification of patients at high risk for breast cancer. For this study, patients were considered at high risk for breast cancer if they had any of the following: a 5-year predicted risk of at least 1.66% by the Gail model,20 a history of lobular carcinoma in situ (LCIS), or a history of breast cancer (T1 or T2, N0) with no evidence of recurrence for the last 2 years. In the calculation of risk, the Gail model includes the variables of current age, number of first-degree relatives with breast cancer, nulliparity or age at first live birth, number of breast biopsies, pathologic diagnosis of atypical hyperplasia, and age at menarche.
We prospectively evaluated 32 breast FNA specimens from 27 women (five women each had two FNA specimens, one from each breast). Woman ranged in age from 39–71 years (mean, 57 years). In all women, random periareolar FNA was performed four times each at the 3 o'clock and 9 o'clock positions in each breast as previously described.2 Briefly, a 1.5-inch, 23-gauge needle attached to a 10-ml syringe was used. The needle was inserted approximately 1–2 cm away from the areola, at 3 o'clock and later at 9 o'clock. Following injection of 2 ml of 1% lidocaine, the aspiration needle was moved in multiple directions to ensure sampling of most of the breast tissue, with emphasis on areas of dense breast tissue, where proliferative glandular tissue is often present. All the FNA samples were pooled in 5 ml of Cytolyte solution. After each aspiration, firm pressure was applied to the aspiration site to prevent hematoma formation. A cold pack was also applied to the breast for approximately 10 min after completion of FNA. Cytologic specimens were prepared using the thin preparation (ThinPrep) technique (Cytyc Corporation, Boxborough, MA, USA). Six to eight slides were prepared from each pooled FNA specimen. One slide was stained with Papanicolaou stain for cytologic diagnosis; the remaining slides were saved in the tissue bank for marker studies per the protocol. Cytologic diagnosis was made on the basis of previously published criteria.4 The categories used were NPE, hyperplasia (with or without atypia), and malignant lesion.
Touch preparation smears from seven cases of invasive carcinoma and nine cases of NPE from women at low risk for breast cancer (women with no history of breast cancer or LCIS or with a Gail model-predicted 5-year breast cancer risk of less than 1.66%) were also included for comparison.
FISH
For FISH, thin preparation slides were processed according to the manufacturer's pretreatment and hybridization protocol. A DNA locus-specific identifier probe complementary for 1p12 (LSI1) and chromosome enumeration probes complementary for chromosome 8-, 11-, and 17-specific pericentromeric repetitive sequences (Vysis Inc., Downers Grove, IL, USA) were directly labeled with SpectrumGold, SpectrumRed, SpectrumGreen, and SpectrumAqua fluorophores, respectively, and combined with human placental blocking DNA in LSI/whole chromosome paint hybridization buffer (Vysis Inc.). Ten microliters of probe-hybridization mix was applied to each slide, and then slides were covered with 22-mm circular cover slips (Fisher Scientific, Pittsburgh, PA, USA). Slides were then placed into a HyBrite Denaturation/Hybridization System (Vysis Inc.) for simultaneous probe/target DNA denaturation at 73°C for 5 min, followed by an overnight hybridization at 37°C. After hybridization, slides were washed in 2 × standard saline citrate (SSC)/0.3% NP-40 at 73°C for 2 min and 2 × SSC/0.3% NP-40 at room temperature for 1 min. After air-drying, slides were mounted with DAPI (4,6-diamidino-2-phenylindole; Vysis Inc.), cover-slipped, and evaluated using an Olympus fluorescence microscope.
FISH Scoring
FISH scoring was performed without knowledge of the cytologic findings according to the method described by King et al.17 Briefly, the entire field was initially viewed using DAPI filter and × 10 microscope objective to confirm the presence of adequate numbers of epithelial cell nuclei. As epithelial cells were largely present in clusters, they were easily distinguished from foam cells, which also retain prominent fluorescent background staining in their cytoplasm. The entire field was next examined using the × 60 objective to scan for the presence of epithelial cell clusters with large and/or irregular nuclei. If present, such nuclei were examined with the appropriate filters for each fluorophore-labeled probe set in a fluorescence microscope according to the manufacturer's instructions (Single Bandpass Filter Sets, Vysis Inc.). The numerical changes in chromosomes 1, 8, 11, and 17 were analyzed in all the clusters of epithelial cells. In individual cells, polysomy was defined as the presence of three or more signals per nucleus, and monosomy was defined the loss of one or both signals per nucleus. Aneusomy was defined as monosomy in >20% of cells or polysomy in >6% of cells. If aneusomy was detected on the initial × 60 scan of the entire field, 60 consecutive large, nonoverlapping epithelial cell nuclei were scored for signal copy number using the appropriate filters for all four probes. If atypical nuclear findings or aneusomy was not detected on the initial × 60 scan of the entire field, 60 consecutive large, nonoverlapping epithelial cell nuclei were scored for signal copy number.
Statistical Analysis
We compared monosomy and polysomy of chromosomes 1, 8, 11, and 17 between patient groups. The percentages of monosomic and polysomic cells were summarized by the median, mean, and range and compared between groups with the Wilcoxon rank sum test.
We considered two combinations of chromosomes—1, 8, and 17 and 1, 8, 11, and 17—because of a previous study by Fehm et al20 in which they indicated that certain chromosomal patterns are associated with malignancy. As previously noted, specimens with polysomy in >6% of cells were considered to be polysomic. We also performed a second analysis in which specimens with polysomy in >3% of cells were considered to be polysomic. If a specimen was polysomic for all chromosomes in a combination, it was considered positive for the combination. The number of patients with polysomy for each combination was compared between patient groups using Fisher's exact test. P values of 0.05 or less were considered statistically significant.
Results
The cytologic findings for the 32 FNA specimens in the high-risk group were NPE in 16 cases and hyperplasia in the other 16 cases. None of the high-risk women had a cytologic finding of ‘malignant lesion’. Generally, aspirates of NPE were mildly cellular, with the epithelial cells arranged in small clusters or flat sheets with uniform, small, rounded nuclei, and moderate to abundant cytoplasm. Apocrine cells and histiocytes were present in some of the cases (Figure 1a). Hyperplastic epithelial changes were moderately cellular, with the epithelial cells forming complex or three-dimentional clusters. Nuclei were enlarged and overlapping, with dense chromatin and an irregular nuclear membrane (Figure 2a).
(a) Papanicolaou ThinPrep preparation of a breast fine-needle aspiration sample showing a group of epithelial cells in a monolayer arrangement interpreted as nonproliferative. (b) Epithelial cells hybridized with centromeric probes for chromosomes 1, 8, 11, and 17 showing diosomy for chromosomes 8 and 17 (1 and 11 are not shown) in the majority of the cells. Fewer than 20% of the cells were monosomic.
(a) Papanicolaou ThinPrep preparation of a breast fine-needle aspiration sample showing a group of epithelial cells with irregular nuclei, nuclear overlap, and hyperchromasia interpreted as hyperplasia. A macrophage is present (upper right). (b) and (c) Clusters and individual epithelial cells hybridized with centromeric probes for chromosomes 1, 8, 11, and 17 showing polysomy for all chromosomes (1 and 11 are not shown in b).
Hybridization of the FISH probes to the DNA revealed changes in chromosome number in 11 (69%) of the 16 cases of NPE and 14 (89%) of the 16 cases of hyperplasia (Figures 1b and 2b and c). Changes in chromosome number were also detected in seven (100%) of the seven carcinoma specimens and none of the nine breast epithelium specimens from low-risk women.
In both the NPE and hyperplasia groups, chromosomal loss was more frequent than chromosomal gain (Table 1). In contrast, in the cancer group, chromosomal gain was more common. The number of chromosomes with polysomy ranged from 0 to 1 in the NPE group, 1–4 (average 2.4) in the hyperplasia group, and 2–4 (average 3) in the cancer group.
In the high-risk group, there were no significant differences in monosomy or polysomy of chromosomes 1, 8, 11, and 17 between patients with NPE and those with hyperplasia (Table 2). On the other hand, high-risk patients had significantly more monosomy of chromosomes 1, 11, and 17 (P=0.001 to 0.004) and significantly more polysomy of chromosome 8 compared to low-risk patients (Table 3). High-risk patients also had significantly less polysomy of chromosomes 1, 8, 11, and 17 (P=0.03–0.001) compared to patients with cancer (Table 4).
No patients had monosomy (monosomy in >20% of cells) for the combination of chromosomes 1, 8, and 17 or for the combination of chromosomes 1, 8, 11, and 17. When polysomy was defined as polysomy in >6% of cells, five patients had polysomy for the combination of chromosomes 1, 8, and 17, and four patients had polysomy for the combination of chromosomes 1, 8, 11, and 17. The proportion of patients with polysomy of chromosomes 1, 8, and 17 combined was significantly higher in the cancer group than in the hyperplasia group (Table 5). When polysomy was defined as polysomy in >3% of cells, seven patients had polysomy for the combination of chromosomes 1, 8, and 17, and five patients had polysomy for the combination of chromosomes 1, 8, 11, and 17. For both combinations, the proportion of patients with polysomy was significantly higher in the cancer group than in the hyperplasia group (Table 5).
Fourteen of the 27 patients had prior biopsy findings of atypical ductal hyperplasia (n=7) or LCIS (n=7). Chromosomal aberrations were detected in nine of the 14 patients with a history of atypical ductal hyperplasia or LCIS and in 12 of the 14 patients with no known history of atypical ductal hyperplasia or LCIS.
Discussion
The development and progression of breast cancer are characterized by the accumulation of numeric changes of many chromosomes and, in particular, by early and/or frequent increases in copy levels of chromosomes 1, 8, 11, and 17. In our series, numerical aberrations of chromosomes 1, 8, 11, and 17 were commonly found in the breast epithelium of women at high risk for breast cancer. High-risk patients had significantly more monosomy in chromosomes 1, 11, and 17 and polysomy in chromosome 8 compared to patients at low risk. These findings are in agreement with findings reported previously9, 10, 11 and suggest that detection of aneusomy may be a useful marker for increased risk of breast cancer development.
In our series, the number of chromosomes with polysomy increased as atypia progressed. No NPE specimens had polysomy in more than one chromosome, whereas the hyperplasia and cancer cases had on average polysomy in 2.4 and 3.0 chromosomes, respectively. Among the high-risk group, however, there were no significant differences in aneusomy between the NPE and hyperplasia groups. The lack of correlation between aneusomy and cell morphology may be partly due to the use of a separate slide preparation for FISH analysis, which prevented direct correlation of aneusomy with cell morphology, or to the fact that high-risk patients harbor chromosomal aberrations irrespective of cell morphology.11
Previous studies suggest that certain patterns of aneusomy are indicative of malignancy.16, 21 In a study by Fehm et al,21 several aneusomic patterns in individual tumor cells were reported to indicate malignancy; the most common were polysomy for chromosomes 1, 8, and 17 (detected in 44% of cases), polysomy for chromosomes 1 and 8 (31% of cases), and polysomy for chromosome 1 (31% of cases). In our study, two of the hyperplastic specimens exhibited polysomy in chromosomes 1, 8, and 17, and this combination of polysomy was significantly more common in the cancer group than in the hyperplasia group. This difference was more significant when the cutoff value for polysomy was lowered to polysomy in >3% of cells. Whether our finding is an early indication of malignancy in these two cases will require follow-up for confirmation. Correlation with other markers of increased risk will also be necessary to determine the biologic significance of these changes.
Both chromosomal gains and chromosomal losses were noted in the high-risk group, while chromosomal gain was the most frequent aberration in the cancer cases. These findings are similar to those reported by others.9, 13, 21 It must be noted that criteria for defining monosomy and polysomy have varied in the literature, making comparison among different series difficult. This problem is compounded when tissue sections rather than cytologic preparations are used because of nuclear truncation, which results in less accurate signal scoring. In a study by Yamamoto et al,19 in which they evaluated ductal lavage specimens, none of the 54 benign lesions (not specified as to cancer risk) demonstrated aneusomy for chromosomes 1, 11, and 17, whereas all six malignant lesions were aneusomic. However, that study used a more stringent definition of aneusomy: polysomy was defined as greater than 20% of nuclei having three or more signals specific for chromosomes 1, 11, and 17, and monosomy was defined as greater than 15% of nuclei having only one or fewer signals specific for chromosomes 1, 11, and 17. Similar findings were reported by Ichikawa et al:16 95.9% of their breast cancer cases demonstrated aneusomy of at least one of the three chromosomes studied, but none of the phyllodes tumors or benign cases showed evidence of aneusomy. Using the same cutoff values for polysomy, only three (10%) of the 32 cases in our high-risk group would be considered polysomic, in addition to 100% of the malignant cases.
Botti et al11 used threshold values of 40% of cells showing signal loss and 13% of cells showing signal gain in their study of tissue imprints of primary tumors and their surrounding uninvolved parenchyma. They found that primary breast tumor and adjacent uninvolved breast parenchyma shared the same pattern of chromosome 1 and 17 aneusomy in 66.7% of patients, and that contralateral benign breast samples were not different from samples of primary breast tumor or adjacent tissue. The results of their study further support the concept that aneusomy is frequently found in normal appearing breast tissue in patients who are at high risk for breast cancer. It remains to be seen, however, whether certain aneusomic patterns are associated with a higher risk of progression to frank malignancy and as a result can be used for predicting progression to a higher-risk lesion.
In summary, our findings indicate that aberrations of chromosomal number are common in women at high risk for breast cancer. There were no significant differences between the NPE and hyperplasia groups with respect to gain or loss of individual chromosomes. However, high-risk cases had significantly more monosomy of chromosomes 1, 11, and 17, and significantly more polysomy of chromosome 8 compared to low-risk cases. Although aneusomy may be a useful marker for separating low-risk from high-risk groups, using aneusomy this way in the clinical setting is impractical, because there are easier and lower cost methods available (eg the Gail model) for risk stratification. However, follow-up studies evaluating the association between specific patterns of chromosomal aneusomy and progression to malignancy would be of interest.
References
Dooley WC, Ljung B-M, Veronesi U, et al. Ductal lavage for detection of cellular atypia in women at high-risk for breast cancer. J Natl Cancer Inst 2001;93:1624–1632.
Fabian CJ, Kimler BF, Zalles CM, et al. Short-term breast cancer prediction by random periareolar fine-needle aspiration cytology and the Gail risk model. J Natl Cancer Inst 2000;92:1217–1227.
Wrensch WR, Petrakis NL, Miike R, et al. Breast cancer risk in women with abnormal cytology in nipple aspirates of breast fluid. J Natl Cancer Inst 2001;93:1791–1798.
The uniform approach to breast fine-needle aspiration biopsy. NIH consensus development conference. Am J Surg 1997;174:371–385.
Sneige N . Fine-needle aspiration cytology of in situ epithelial cell proliferation in the breast. Am J Clin Pathol 2000;113 (Suppl 1):S38–S48.
Sneige N . Ductal lavage for breast cancer risk assessment. Patholo Case Rev 2003;8:233–239.
Harrison MM, Magee HM, Loughlin JFO, et al. Comparison of chromosome 1 aneusomy detected by interphase cytogenetics and DNA ploidy in carcinoma of the breast. Histopathology 1997;30:221–226.
Mendelin J, Grayson M, Wallis T, et al. Analysis of chromosome in situ aneuploidy in breast carcinoma progression by using fluorescent in situ hybridization. Lab Invest 1999;79:387–393.
Sneige N, Sahin A, Dinh M, et al. Interphase cytogenetics in mammographically detected breast lesions. Hum Pathol 1996;27:330–335.
Illei PB, Feiner HD, Symmans WF, et al. Numerical abnormalities of chromosomes 7, 18, and X in precancerous breast disease defined by fluorescent in situ hybridization. Breast J 1998;4:252–257.
Botti C, Pescatore B, Mottolese M, et al. Incidence of chromosomes 1 and 17 aneusomy in breast cancer and adjacent tissue: an interphase cytogenetic study. J Am Coll Surg 2000;190:530–539.
Visscher D, Jimenez RE, Grayson M, et al. Histopathologic analysis of chromosome aneuploidy in ductal carcinoma in situ. Hum Pathol 2000;31:201–207.
Marinho AF, Botelho M, Schmitt FC . Evaluation of numerical abnormalities of chromosomes 1 and 17 in proliferative epithelial breast lesions using fluorescence in situ hybridization. Pathol Res Pract 2000;196:227–233.
Cummings MC, Aubele M, Mattis A, et al. Increasing chromosome 1 copy number parallels histological progression in breast carcinogenesis. Br J Cancer 2000;82:1204–1210.
Micale MA, Visscher DW, Gulino SE . Chromosomal aneuploidy in proliferative breast disease. Hum Pathol 1994;25:29–35.
Ichikawa D, Hashimoto N, Hoshima M, et al. Analysis of numerical aberrations of specific chromosomes by fluorescent in situ hybridization as a diagnostic tool in breast cancer. Cancer 1996;15:2064–2069.
King BL, Tsai SC, Gryga ME, et al. Detection of chromosomal instability in paired breast surgery and ductal lavage specimens by interphase fluorescence in situ hybridization. Clinical Cancer Res 2003;9:1509–1516.
Krishnamurthy S, Zhao L, Hayes K, et al. Feasibility and utility of using chromosomal aneusomy to further define the cytologic categories in nipple aspirate fluid specimens. Cancer 2004;102:322–327.
Yamamoto D, Senzaki H, Nakagawa H, et al. Detection of chromosomal aneusomy by fluorescence in situ hybridization for patients with nipple discharge. Cancer 2003;97:690–694.
Gail MH, Brinton LA, Byar DP, et al. Projecting individualized probabilities of developing breast cancer for white females who are being examined annually. J Natl Cancer Inst 1989;81:1879–1886.
Fehm T, Morrison L, Saboorian H, et al. Patterns of aneusomy for three chromosomes in individual cells from breast cancer tumors. Breast Cancer Res Treat 2002;75:227–239.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sneige, N., Liu, B., Yin, G. et al. Correlation of cytologic findings and chromosomal instability detected by fluorescence in situ hybridization in breast fine-needle aspiration specimens from women at high risk for breast cancer. Mod Pathol 19, 622–629 (2006). https://doi.org/10.1038/modpathol.3800571
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.3800571