Abstract
Basal-like carcinomas have recently been identified in gene expression profiling studies as a subtype of invasive breast cancer. These lesions are estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative (triple negative), and typically express basal cytokeratins, epidermal growth factor receptor (EGFR), and/or c-kit. As poorly differentiated invasive ductal carcinomas, they presumably have a ductal carcinoma in situ (DCIS) precursor with similar cytologic and immunophenotypic features. However, the frequency and even the existence of a DCIS lesion with an immunophenotype analogous to that of invasive basal-like carcinomas have not been previously evaluated. We studied 66 cases of high nuclear grade DCIS using antibodies to ER, PR, HER2, three basal cytokeratins, EGFR, and c-kit to determine the frequency of the triple negative phenotype, and to determine the relationship between the triple negative phenotype and expression of basal cytokeratins and other biomarkers characteristically expressed by invasive basal-like carcinomas. Four cases (6%) exhibited the triple negative phenotype; the remaining cases showed other combinations of ER, PR, and HER2 expression (nontriple negative). Basal cytokeratins, EGFR, or both were expressed by all four triple negative lesions, but by only 21 of 51 (42%) nontriple negative cases (P=0.04). We conclude that a small proportion of high-grade ductal carcinomas in situ exhibit an ER-negative/PR-negative/HER2-negative (triple negative) phenotype, and these lesions more commonly show expression of basal cytokeratins and/or EGFR than nontriple negative high-grade DCIS. Given that invasive breast cancers typically share immunophenotypic features with the ductal carcinoma in situ from which they arise, our findings raise the possibility that the triple-negative, basal cytokeratin and/or EGFR-positive DCIS lesions we identified represent a precursor lesion to invasive basal-like carcinomas.
Similar content being viewed by others
Main
Recent gene expression profiling studies have identified a subtype of breast cancer known as basal-like carcinomas.1, 2, 3, 4 These lesions constitute about 15% of invasive breast cancers, are characterized by lack of expression of estrogen receptor (ER) and progesterone receptor (PR), and absence of HER2 protein overexpression (the so-called ‘triple negative’ phenotype), and have a poor prognosis.5, 6, 7, 8 Basal-like carcinomas typically exhibit expression of one or more of the basal cytokeratins (such as CK5/6, CK14, and CK17), epidermal growth factor receptor (EGFR), and/or c-kit.5, 6, 7, 8 These tumors are often seen in women with BRCA1 mutations, but also occur among sporadic breast cancers.4, 5, 6, 7, 9 In expression array studies, the majority of triple negative invasive breast cancers cluster with basal-like carcinoma.4, 5, 6, 7, 10
Basal-like carcinomas are poorly differentiated invasive ductal carcinomas,4, 5, 7 and presumably have a ductal carcinoma in situ (DCIS) precursor with similar cytologic and immunophenotypic features. However, the frequency and even the existence of a distinctive DCIS lesion with an immunophenotype similar to that of basal-like carcinoma has not been previously identified.
To address this issue, we studied 66 cases of high nuclear grade DCIS to determine the frequency of the triple negative phenotype, and to determine the relationship between the triple negative phenotype and expression of basal cytokeratins and other biomarkers characteristically expressed by invasive basal-like carcinomas.
Materials and methods
We identified 66 cases of high nuclear grade DCIS accessioned in the Pathology Department of Beth Israel Deaconess Medical Center, Boston, MA, USA between 2000 and 2005 for which paraffin blocks were available, and for which there was sufficient DCIS remaining in the blocks to permit the cutting of additional sections for immunohistochemical staining. Forty-four cases were excisional biopsies, 17 were mastectomy specimens, and five were core needle biopsies. Fifty cases were composed of DCIS alone. In the remaining 16 cases, the DCIS analyzed was from specimens that also contained invasive breast cancer, but the blocks selected for study contained only DCIS, without an invasive component. For each case, all available hematoxylin and eosin-stained sections were reviewed to confirm the diagnosis of high-grade DCIS and to select a representative block for immunostaining. DCIS nuclear grade was determined using the nuclear grading criteria of Lagios.11, 12
We performed immunohistochemical staining for ER, PR, HER2, three basal cytokeratins (CK5/6, CK14 and CK17), EGFR, and c-kit on sections cut from one representative paraffin block from each case. These particular biomarkers were selected for analysis because they are the markers that have been most commonly used to identify invasive basal-like breast cancers.5, 6, 7, 8 Sources and dilutions of the primary antibodies used in this study are listed in Table 1. Immunostains for ER were performed using a method described in detail previously.13 Immunostains for PR, HER2, CK5/6, CK14, and CK17 were performed on tissue sections following deparaffinization in two 5-min changes of xylene and rehydration through graded alcohols to distilled water. After blocking endogenous peroxidase activity, sections were subjected to heat-induced epitope retrieval by heating in a vegetable steamer in citrate buffer (pH 6.0) for 30 min. Following heat-induced epitope retrieval, the primary monoclonal antibodies were applied to the sections for 30 min at room temperature. The slides were then incubated with HRP Labeled Polymer (DakoCytomation). The reactions were completed with the Envision detection system (DakoCytomation) using 3-3′ diaminobenzidine as the chromogen.
After the staining runs for ER, PR, HER2, and the three basal cytokeratins had been completed, 55 cases had sufficient DCIS remaining in the block for EGFR immunostains and 54 cases had sufficient material for c-kit immunostains. Immunostaining for EGFR was performed on a DakoCytomation Autostainer using the EGFR pharmDx Kit (DakoCytomation). Antigen unmasking for EGFR was performed by proteolytic enzyme digestion (Proteinase K for 5 min). The immunostaining procedure used for c-kit was as described above for PR, HER2, and the basal cytokeratins.
Positive controls were included in each staining run and consisted of breast cancers known to express each of the antigens of interest. Negative controls in which the primary monoclonal antibodies were replaced by mouse myeloma protein were performed for each case.
Each immunostained slide was evaluated for the presence of ER and PR expression, HER2 protein overexpression, and expression of CK5/6, CK14, CK17, EGFR, and c-kit in the DCIS cells. Tumor cells that showed nuclear staining for ER or PR were considered ER-positive or PR-positive, respectively. Of note, all ER-positive and PR-positive cases showed staining in at least 10% of the DCIS tumor cell nuclei, whereas all ER-negative cases showed complete absence of tumor cell staining (but with staining of normal breast epithelial cell nuclei). Tumor cells were considered positive for HER2 protein over-expression when greater than 10% of the cells showed strong membrane staining (equivalent to a score of 3+ in the DakoCytomation HercepTest). Cases were considered basal cytokeratin-positive, EGFR-positive, and c-kit-positive if any cytoplasmic and/or membranous staining was observed in the DCIS cells, even if focal. These criteria are similar to those previously used for scoring these markers in invasive basal-like cancers.5 All immunostains were initially reviewed and scored by one of the study pathologists (BB) with a second, simultaneous review of the basal cytokeratin, EGFR, and c-kit immunostains by the other two pathologists.
Fisher's exact test was used to compute the statistical association between two variables and the Mann–Whitney test was used to compare means. A P-value of ⩽0.05 was considered to be statistically significant.
The study was approved by the Beth Israel Deaconess Medical Center Committee on Clinical Investigations.
Results
Among the 66 cases of high-grade DCIS studied, all patients were women and the median age was 51 years (range 31–89 years). The frequency of expression of the various biomarkers studied is summarized in Table 2. Overall 56% of the cases were ER positive, 40% were PR positive, and 67% showed HER2 protein overexpression. Four cases (6%) were ER negative/PR negative/HER2 negative (ie triple negative) and the remaining 59 cases with sufficient tissue remaining showed other combinations of ER, PR, and HER2 expression (nontriple negative), including 12 ER+/PR+/HER2+, 12 ER+/PR+/HER2−, 24 ER−/PR−/HER2+, 6 ER+/PR−/HER2−, and 5 ER+/PR−/HER2+. No DCIS was present on the sections immunostained for PR in three cases. For the purposes of analysis, these cases were included in the nontriple negative group, since each was either ER positive (n=2) or showed HER2 overexpression (n=1).
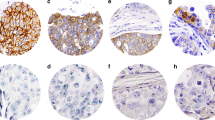
Staining for one or more of the basal cytokeratins was detected in 21 of the 66 cases (32%). A total of 14 were CK5/6+/CK14+/CK17+, two were CK5/6+/CK14−/CK17+, two were CK5/6−/CK14+/CK17+, and three were CK17+ only. In all of these cases, basal cytokeratin staining was focal, consistent with the distribution of basal cytokeratin expression previously reported in invasive basal-like carcinoma8 (Figure 1). Of the four triple negative cases, three (75%) were basal cytokeratin-positive, whereas only 18 of the 62 (29%) nontriple-negative DCIS cases were basal cytokeratin-positive (P=0.09).
‘Triple negative’ high-grade ductal carcinoma in situ. (a) This case of high grade ductal carcinoma in situ exhibited the ‘triple negative’ phenotype (estrogen receptor negative, progesterone receptor negative, and negative for HER2 protein overexpression). (b) Immunostain for basal cytokeratin CK5/6 demonstrates variable staining of the neoplastic cells.
EGFR expression was detected in 12 of 55 evaluable cases (22%) and was significantly more common among the triple negative (three of four, 75%) than among the nontriple negative high-grade DCIS (nine of 51, 18%) (P=0.03). Either basal cytokeratins, EGFR, or both were expressed by all four triple negative high-grade DCIS (100%), but by only 21 of 51 (41%) nontriple negative cases (P=0.04). c-kit expression was identified in only two (one triple negative and one nontriple negative DCIS).
The details of the immunostaining patterns of the four triple-negative DCIS cases identified in this study are summarized in Table 3. Of note, these four cases did not exhibit any histologic features that distinguished them from the nontriple negative high-grade DCIS cases in this population. While patients with triple negative DCIS were on average older than those with nontriple negative lesions (mean age 67 vs 53 years, respectively), this difference was not statistically significant (P=0.11).
Discussion
Recent studies using cDNA microarray technology and unsupervised cluster analysis have provided new insights into the classification of invasive breast cancers.2, 3, 4, 14 These studies have resulted in the identification of several breast cancer subgroups that differ in their gene expression signatures and clinical course. The molecularly distinct breast cancer subgroups identified to date include luminal subtypes A and B (both of which are hormone receptor positive), the HER2 subtype, and a group of breast cancers not previously recognized as a distinct subset, the basal-like cancers.2, 3, 4, 14
Histopathologic studies of basal-like carcinomas have indicated that these tumors are poorly differentiated invasive ductal carcinomas and typically have little or no associated DCIS.4, 5, 7 Breast carcinomas described prior to the era of the gene expression profiling studies as poorly differentiated invasive ductal carcinomas with myoepithelial differentiation and central acellular zones,15 or with fibrotic foci16 most likely belong in this group.
It is reasonable to speculate that, like other invasive ductal carcinomas, basal-like carcinomas arise from a DCIS precursor and that such a precursor might share immunophenotypic features with the subsequent invasive cancer. However, the frequency and even the existence of a distinctive DCIS lesion with an immunophenotype analogous to that of basal-like carcinoma has not been previously identified.
To test the hypothesis that there may be a subset of DCIS lesions that has immunophenotypic features similar to invasive basal-like carcinoma and that may represent a basal-like carcinoma precursor, we examined a series of cases of high-grade DCIS to assess the frequency of expression of a panel of biomarkers that characterize invasive basal-like carcinomas. The study population was restricted to DCIS lesions of high nuclear grade because in our experience, low and intermediate nuclear grade DCIS lesions are invariably ER positive,17 precluding their possible classification as ‘triple negative.’
We found that among 66 high grade DCIS cases, a small proportion (four cases, 6%) exhibited the ER negative/PR negative/HER2 negative phenotype that characterizes invasive basal-like carcinomas. Moreover, similar to invasive basal-like carcinoma, all four of these triple negative high-grade DCIS showed expression of one or more basal cytokeratins and/or EGFR. There were no histologic features that distinguished the triple negative from the nontriple negative high-grade DCIS in this series. Of interest, even among the small number of triple negative DCIS identified in this study, there was immunophenotypic heterogeneity with regard to basal cytokeratin, EGFR, and c-kit expression (Table 3). This is similar to the heterogeneity in expression of these biomarkers previously reported in a series of invasive basal-like cancers defined by gene expression profiling.5
The results provided by this observational study are clearly not sufficient to prove a precursor–product relationship, particularly in view of the small number of cases of triple negative DCIS. However, our findings raise the interesting possibility that the triple negative/basal cytokeratin and/or EGFR positive high-grade DCIS we have identified may represent a precursor lesion of invasive basal-like carcinomas. The infrequent occurrence of DCIS lesions with the triple negative, basal-like immunophenotype, in conjunction with observations from prior studies that have demonstrated that invasive basal-like carcinomas usually have little or no identifiable component of DCIS, could be explained by the fact that basal-like carcinomas evolve rapidly and largely obliterate the DCIS from which they arise.
References
Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature 2000;406:747–752.
Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA 2001;98:10869–10874.
Sorlie T, Tibshirani R, Parker J, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA 2003;100:8418–8423.
Brenton JD, Carey LA, Ahmed AA, et al. Molecular classification and molecular forecasting of breast cancer: ready for clinical application? J Clin Oncol 2005;23:7350–7360.
Nielsen TO, Hsu FD, Jensen K, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res 2004;10:5367–5374.
Abd El-Rehim DM, Pinder SE, Paish CE, et al. Expression of luminal and basal cytokeratins in human breast carcinoma. J Pathol 2004;203:661–671.
Abd El-Rehim DM, Ball G, Pinder SE, et al. High-throughput protein expression analysis using tissue microarray technology of a large well-characterised series identifies biologically distinct classes of breast cancer confirming recent cDNA expression analyses. Int J Cancer 2005;116:340–350.
van de Rijn M, Perou CM, Tibshirani R, et al. Expression of cytokeratins 17 and 5 identifies a group of breast carcinomas with poor clinical outcome. Am J Pathol 2002;161:1991–1996.
Foulkes WD, Stefansson IM, Chappuis PO, et al. Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J Natl Cancer Inst 2003;95:1482–1485.
Hedenfalk I, Duggan D, Chen Y, et al. Gene-expression profiles in hereditary breast cancer. N Engl J Med 2001;344:539–548.
Lagios MD, Margolin FR, Westdahl PR, et al. Mammographically detected duct carcinoma in situ. Frequency of local recurrence following tylectomy and prognostic effect of nuclear grade on local recurrence. Cancer 1989;63:618–624.
Scott MA, Lagios MD, Axelsson K, et al. Ductal carcinoma in situ of the breast: reproducibility of histological subtype analysis [see comments]. Hum Pathol 1997;28:967–973.
Collins LC, Botero ML, Schnitt SJ . Bimodal frequency distribution of estrogen receptor immunohistochemical staining results in breast cancer: an analysis of 825 cases. Am J Clin Pathol 2005;123:16–20.
Perou CM, Jeffrey SS, van de Rijn M, et al. Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proc Natl Acad Sci USA 1999;96:9212–9217.
Tsuda H, Takarabe T, Hasegawa T, et al. Myoepithelial differentiation in high-grade invasive ductal carcinomas with large central acellular zones. Hum Pathol 1999;30:1134–1139.
Hasebe T, Sasaki S, Imoto S, et al. Prognostic significance of fibrotic focus in invasive ductal carcinoma of the breast: a prospective observational study. Mod Pathol 2002;15:502–516.
Collins LC, Schnitt SJ . HER2 protein overexpression in estrogen receptor-positive ductal carcinoma in situ of the breast: frequency and implications for tamoxifen therapy. Mod Pathol 2005;18:615–620.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bryan, B., Schnitt, S. & Collins, L. Ductal carcinoma in situ with basal-like phenotype: a possible precursor to invasive basal-like breast cancer. Mod Pathol 19, 617–621 (2006). https://doi.org/10.1038/modpathol.3800570
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.3800570
Keywords
This article is cited by
-
Secretory breast carcinoma in a female adult with liver metastsis: a case report and literature review
Diagnostic Pathology (2021)
-
Triple-negative and HER2 positive ductal carcinoma in situ of the breast: characteristics, behavior, and biomarker profile
Virchows Archiv (2018)
-
Comparison of HER2 amplification status among breast cancer subgroups offers new insights in pathways of breast cancer progression
Virchows Archiv (2017)
-
Breast cancer complexity: implications of intratumoral heterogeneity in clinical management
Cancer and Metastasis Reviews (2017)
-
Ductal carcinoma in situ of the breast: correlation between histopathological features and age of patients
Diagnostic Pathology (2014)