Abstract
Dedifferentiated liposarcoma is a malignant adipocytic neoplasm containing a nonlipogenic sarcoma of variable histological grade that arises against the background of a pre-existing well-differentiated liposarcoma. The phenomenon of dedifferentiation is considered to be time-dependent, but the mechanism is not well known. The retinoblastoma protein, encoded by the RB1 gene located at 13q14, is a key regulator of proliferation, development, and differentiation of certain cell types, including adipocytes. In the current study, we investigated the genetic alterations of the RB1 gene, such as mutation (the essential promoter region and the protein-binding pocket domain; exons 20–24) and methylation of the promoter region, in addition to pRB expression and loss of heterozygosity (LOH) status, in two morphologically distinct areas (nonlipogenic dedifferentiated and well-differentiated components) in 27 patients. As a control, 11 undifferentiated high-grade pleomorphic sarcoma/pleomorphic malignant fibrous histiocytoma samples and 11 well-differentiated liposarcoma samples were also evaluated. Dedifferentiated components showed LOH (15/25; 60%) and abnormal retinoblastoma protein expression (18/27; 66.7%) more frequently than noted in the well-differentiated components (3/24; 12.5% and 9/27; 33.3%, respectively). Five and four out of the 27 dedifferentiated components harbored mutations and promoter methylation, respectively, whereas none of these alterations were seen in the well-differentiated components. These results suggest that retinoblastoma protein has a major role to play in dedifferentiation and that a ‘two-hit’ mechanism is involved in the altered retinoblastoma protein expression in dedifferentiated liposarcoma.
Similar content being viewed by others
Main
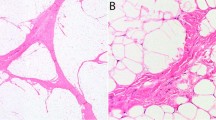
Dedifferentiated liposarcoma is defined as a neoplasm with a well-differentiated liposarcoma juxtaposed against areas of high-grade non-lipogenic sarcoma, or as a high-grade non-lipogenic sarcoma arising at precisely the same location that had previously been the primary site of well-differentiated liposarcoma (Figure 1a–c). Most of the dedifferentiated liposarcomas display extensive areas of high-grade dedifferentiation resembling malignant fibrous histiocytoma or high-grade fibrosarcoma, whereas some cases contain areas of only low-grade dedifferentiation.1, 2 The behavior of dedifferentiated liposarcoma is more aggressive than that of pure well-differentiated liposarcoma. Dedifferentiation is mostly considered a time-dependent phenomenon, and several reports have suggested an association between dedifferentiation and the altered expression of specific proteins, such as MDM2, P53, H-ras, β-catenin, and retinoblastoma protein (pRB).3, 4, 5, 6, 7
pRB negatively regulates the cellular G1/S transition of the proliferative cell cycle and is required for proper differentiation of certain cell types, including skeletal muscle and adipocytes.8 Decreased expression of pRB in malignant mesenchymal tumors has been reported by some authors based on immunohistochemistry or Western blotting.9, 10, 11, 12 The retinoblastoma gene (RB1) at chromosome 13q14 was originally identified as the gene responsible for the development of retinoblastoma and has served as a prototype of human tumor suppressor genes. Biallelic inactivation of RB1 is a hallmark not only of retinoblastoma, but has also been described in a variety of other tumors, including osteosarcoma and malignant fibrous histiocytoma.13, 14 Schneider-Stock et al analyzed RB1-loss of heterozygosity (LOH) in 11 dedifferentiated liposarcoma patients and concluded that RB1-LOH plays an important role in the tumor progression of well-differentiated liposarcoma to dedifferentiated liposarcoma;7 however, whether or not there are alterations to the other allele in this tumor has not been previously ascertained. In the present study, we first performed LOH analysis using five microsatellite markers at 13q12–q14 in 27 cases of dedifferentiated liposarcoma. Then, mutation and methylation analysis screening for genetic changes of the RB1 gene was performed, in addition to immunohistochemical analysis.
Materials and methods
Patients and DNA Extraction
In all, 27 patients with dedifferentiated liposarcoma were included in this study. As a control, 11 cases of well-differentiated liposarcoma and 11 cases of malignant fibrous histiocytoma were used. Recently, some authors have suggested that most retroperitoneal malignant fibrous histiocytomas have the possibility of actually being dedifferentiated liposarcomas.15, 16 Accordingly, we selected those cases of malignant fibrous histiocytoma where the tumors arose only in the extremities or the trunk. The diagnosis of all the malignant fibrous histiocytomas was confirmed by the panels of immunohistochemical study, since malignant fibrous histiocytoma is defined as an undifferentiated pleomorphic sarcoma which shows no distinct line of differentiation. All the cases were collected from among the soft-tissue sarcomas that had been registered in the Department of Anatomic Pathology, Graduate School of Medical Sciences, Kyushu University, Japan, between 1982 and 2004. In all, 12 cases of dedifferentiated liposarcoma had been examined in previous mutation analyses.5, 6 All the materials were fixed in 10% formaldehyde and then embedded in paraffin. Separate blocks from two morphologically distinct components of dedifferentiated liposarcoma were selected for both immunohistochemical and molecular studies. Clinicopathological data of the 27 patients with dedifferentiated liposarcoma, the 11 patients with well-differentiated liposarcoma, and the 11 patients with malignant fibrous histiocytoma are listed in Tables 1 and 2, respectively. The clinical data of these patients were obtained from their medical records.
Genomic DNA was isolated from all the cases using standard proteinase K digestion and phenol/chloroform extraction, and was used for the following molecular analysis.
LOH Analysis
Tumor and normal DNA samples were subjected to PCR using primers for the following five dinucleotide microsatellite markers at 13q12–14: D13S175, D13S153, D13S233, D13S1293, and D13S1312. D13S153 is located within intron 2 of the RB1 gene. Primer sequences were obtained by use of the NCBI UniSTS database (http://www.ncbi.nlm.nih.gov/). The forward primer was end-labeled with 6-carboxyfluorescein (6-FAM) at the 5′ end. The procedure was carried out according to a method described previously.17, 18 The data which were processed using GeneScan software (Applied Biosystems) were compared between the tumor and normal DNA for each patient. Informative cases were defined as when the heterozygous alleles were identified within normal DNA. LOH was defined as when an allelic imbalance was observed (the detected allele of the tumor DNA was less than 50% of that of the corresponding normal DNA). Reproducibility was confirmed by 2–4 independent PCR amplifications for each sample.
Mutation Analysis by PCR-SSCP
Mutation analysis was performed for the essential promoter region (encompassing nucleotides −300 to −174) and for the protein-binding pocket domain (exons 20–24) of the RB1 gene. PCR was performed in a final reaction volume of 20 μl containing 100 ng of template DNA, 1.5 mM MgCl2, 1 × PCR buffer (Applied Biosystems, Foster City, CA, USA), 0.25 mM of dNTP mix, 0.5 μM each of sense and antisense primer, and 1 U of Gold Taq polymerase (Applied Biosystems). DNA sequences were amplified for 40 cycles. Primer sequences and annealing temperature are listed in Table 3. Human genomic DNA (Clontech, Palo Alto, CA, USA) was used as a positive control for each PCR and for all the subsequent reactions. SSCP was performed using a gel containing 12.5% acrylamide (GenePhor™, Amersham Pharmacia Biotech, Uppsala, Sweden) and DNA fragment analyzer (GenePhor™, Amersham Pharmacia Biotech), and then the DNA bands were visualized by a DNA Silver Staining Kit (GenePhor™, Amersham Pharmacia Biotech). To increase the quantity of mutant DNA prior to sequencing, the extra bands that seemed to be aberrantly migrating were excised from the SSCP gel and re-amplified using the same primers. The sequence data were obtained using ABI Prism 310 Collection Software, and were analyzed using Sequencing Analysis and Sequence Navigator Software.
Methylation-Specific PCR for the Promoter Region of the RB1 Gene
Bisulfite conversion was performed with 1 μg of genomic DNA and the reagents provided with Intergen's CpGenome DNA Modification Kit (Intergen, New York, NY, USA). Methylation-specific PCR was performed to determine the DNA methylation status of CpG islands of the promoter region of the RB1 gene. Primer pairs were designed according to criteria described previously.19 The composition of PCR mixes was the same as that of the mutation analysis described above. PCR of bisulfite-treated template DNA was carried out for 35 cycles. To ensure PCR amplification of the methylated RB1 promoter sequence after modification, we methylated genomic DNA in vitro with the CpG methylase enzyme Sss-I (New England BioLabs, Beverly, MA, USA). This DNA was then subjected to sodium bisulfite modification as described above and served as a positive control. In addition, DNA of normal skeletal muscle was used as a negative control.
pRB Immunohistochemistry
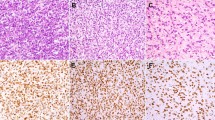
Immunohistochemistry was performed using the anti-pRB mouse monoclonal antibody (clone G3–245, which recognizes RB1 exons 9–12; PharMingen, San Diego, CA, USA; 1:1000). According to the previously published criteria,20 the intensity and pattern of pRB nuclear staining were used to separate the cases into one of three groups. Group 1 comprised patients whose tumors had minimal or undetectable nuclear staining (<20% of tumor cells), and these were considered to be pRB-negative. Patients whose tumors were stained in a heterogeneous pattern (20–80% of tumor cells) were classified as Group 2. The staining of Group 3 was strongly positive with a homogenous pRB nuclear immunoreactivity (>80% of tumor cells). Vascular endothelial cells in each specimen were used as an internal positive control.
Statistical Analysis
We performed Fisher's exact test to assess the correlation among various factors. A P-value of less than 0.05 was considered to be statistically significant.
Results
Clinical Findings
The distribution of clinicopathologic characteristics is outlined in Table 1. Patients with dedifferentiated liposarcoma ranged from 28 to 82 years of age (mean, 59.7 years), whereas those with well-differentiated liposarcoma and malignant fibrous histiocytoma ranged from 41–79 years (mean, 59.7 years) and from 52–76 years (mean, 63.8 years), respectively. Dedifferentiated liposarcoma showed a male predominance of 20 men to seven women, whereas well-differentiated liposarcoma was almost evenly distributed. In all, 18 cases of dedifferentiated liposarcoma occurred in the retroperitoneum, one case each occurred in the mediastinum and mesenterium, and the others occurred in accessible sites, including the thigh (three cases), abdominal wall (one case), back (one case), groin (one case), and lower leg (one case). Histologically, 23 cases of dedifferentiated component showed a proliferation of atypical spindle-shaped and pleomorphic cells arranged in fascicles or in a storiform pattern, mimicking storiform-pleomorphic type malignant fibrous histiocytoma. Two and one cases had the pathologic features of myxoid malignant fibrous histiocytoma and inflammatory malignant fibrous histiocytoma, respectively. One case consisted of elongated slender spindle-shaped cells of a uniform appearance surrounded by abundant collagen, resembling fibromatosis (Case D27, Figure 2). In all, 23 cases were de novo tumors, whereas the others showed secondary dedifferentiation.
LOH on Chromosome 13q12–14
As shown in Table 4, LOH analysis at the 13q12–14 locus was performed in 27 patients. Two cases showed noninformative findings for all five markers. LOH for one or more markers was found in 15 out of 25 (60%) cases with dedifferentiated component and in three out of 24 (12.5%) cases with well-differentiated component. Representative examples of LOH are depicted in Figure 3. The frequency of LOH in dedifferentiated components was significantly higher than that in well-differentiated components (P=0.0009). All the LOH-positive well-differentiated components also demonstrated LOH positivity with concomitant dedifferentiated components. Most cases with dedifferentiated component showed LOH from 13q12 to 13q14, while only one case with well-differentiated component showed LOH at two or more markers (Case D17). Among the control cases, LOH for one or more markers was found in six out of 11 malignant fibrous histiocytomas (54.5%), but in only one out of 11 well-differentiated liposarcomas (9.1%) (Table 2).
Mutation of Exons 20–24 and the Promoter Region of the RB1 Gene
The results of mutational analysis are summarized in Table 4. SSCP analysis followed by DNA direct sequencing revealed five missense mutations in 27 dedifferentiated components (18.5%, Figure 4a–b). None of the cases with well-differentiated component showed any sequence changes, and none of the cases had mutations within the essential promoter region. In four out of the five cases with RB1 mutation, we detected LOH at the RB1 intragenic marker (D13S153). The remaining one case with RB1 mutation was not informative at D13S153, although this case did show LOH for the other markers (Case D24).
RB1 Promoter Hypermethylation
Methylated and unmethylated control DNA showed the expected fragment sizes of 78 and 83 bp, respectively. RB1 promoter hypermethylation was detected in four out of 27 (14.8%) dedifferentiated components, whereas no hypermethylation was detected in any of the well-differentiated components (Figure 5). In the control cases, promoter hypermethylation was detected in four out of 11 (36.4%) malignant fibrous histiocytomas. No hypermethylation was detected in any of the well-differentiated liposarcomas.
pRB Expression
Immunohistochemical results in dedifferentiated liposarcoma are listed in Table 4. Alterations in pRB expression were observed in 18 of the dedifferentiated components (66.7%) and in nine of the well-differentiated components (33.3%) among the 27 patients, on the basis of minimal or heterogeneous staining (Groups 1 and 2, Figure 6a–b). In all, 15 LOH-positive dedifferentiated components showed significantly decreased expression (Groups 1 and 2, P=0.009). This correlation was not observed in well-differentiated component. All the dedifferentiated components with RB1 mutation and four out of the five dedifferentiated components with RB1 promoter hypermethylation showed decreased expression.
Discussion
Dedifferentiated liposarcoma is one of the subtypes of liposarcoma, which is the most common soft-tissue sarcoma in adults, and it occurs in the retroperitoneum, the abdominal cavity, and the lower extremities. The term ‘dedifferentiation’ is defined as the abrupt transition of a low-grade, well-differentiated sarcoma to high-grade morphology, mostly resembling malignant fibrous histiocytoma either in the primary tumor (de novo) or in local recurrence; this seems to occur in well-differentiated liposarcoma in about 12% of the cases.21 Although most of the dedifferentiated liposarcomas display extensive areas of high-grade dedifferentiation, some cases have been described as having only areas of low-grade dedifferentiation resembling fibromatosis or well-differentiated fibrosarcoma.1, 2 In this study, one case showed low-grade dedifferentiation. The behavior of dedifferentiated liposarcoma is more aggressive than that of pure well-differentiated liposarcoma, with a local recurrence rate of 41–75%, a distant metastasis rate of 9–20%, and a disease-related mortality rate of 30–50%,1, 2, 22 and low-grade dedifferentiation is not associated with an improved outcome.1
Cytogenetic, CGH, FISH, and microarray analyses have revealed supernumerary ring or giant marker chromosomes containing amplified DNA sequences in the 12q13–15 in dedifferentiated liposarcoma identical to those detected in well-differentiated liposarcoma.23, 24, 25, 26 It is therefore suggested that well-differentiated liposarcoma and dedifferentiated liposarcoma compromise one subgroup with a broad spectrum in morphology and biologic behavior; however, the mechanism of dedifferentiation is not well known. Chibon et al and Coindre et al have recently demonstrated that most of the malignant fibrous histiocytomas which develop in the retroperitoneum have these alterations.15, 16 Dedifferentiation is mostly considered a time-dependent phenomenon, and several reports have suggested the association between dedifferentiation and the altered expression of specific proteins. Overexpression of the proteins including MDM2 and CDK4, which are encoded by the genes located in the 12q13–15 region, is well described in liposarcoma, and dedifferentiated components exhibit a high level of MDM2-positive inmmunoreactivity in dedifferentiated liposarcomas.3, 4 Hostein et al investigated the amplification level of MDM2 and CDK4 using quantitative real-time PCR, and higher-level amplification was observed in dedifferentiated liposarcomas than in well-differentiated liposarcomas.27 Other than these genes, Sakamoto et al suggested that alteration of the β-catenin and H-ras gene is involved in the dedifferentiation in dedifferentiated liposarcoma.5, 6 Schneider-Stock et al investigated RB1-LOH in two microdissected components of 11 dedifferentiated liposarcoma patients, and RB1-LOH was detected in all the dedifferentiated components, using four intragenic RB1 markers and restriction fragments analysis.7 In our present study, LOH for one or more markers out of five microsatellite markers at 13q12–14 was found in 15 out of 25 (60%) cases with dedifferentiated component and in three out of 24 (12.5%) cases with well-differentiated component. We detected LOH at D13S153, one of the five microsatellite markers located within intron 2 of the RB1 gene, in 13 out of 24 (54%) dedifferentiated components and in two out of 23 (8.7%) well-differentiated components. LOH was more frequently found in dedifferentiated components than in well-differentiated components, and this result is consistent with that of Schneider-Stock et al, although the LOH rate is different (54 vs 100% in dedifferentiated component and 8.7 vs 0% in well-differentiated component). This difference may be explained by the number of cases, different primers, different analyzing procedures, or a different judging standard of LOH. Most dedifferentiated components showed a wide range of LOH from 13q12 to q14. Although it has not been ruled out that other genes in the region of 13q, such as BRCA2, are involved in the tumor progression of well-differentiated liposarcoma to dedifferentiated liposarcoma, in the current study we investigated the alteration of the RB1 gene, one of the best characterized tumor suppressor genes.
The RB1 gene, located on the long arm of chromosome 13, is one of the best-characterized tumor-suppressor genes, and its inactivation has been noted in a variety of human tumors. Biallelic inactivation of RB1 is a hallmark not only of retinoblastoma, but has also been described in a variety of other tumors. Chibon et al reported that RB1 mutations and/or homozygous deletions were found in seven out of 34 malignant fibrous histiocytomas, in addition to frequent (78%) losses of the 13q14–q21 region.14 In the present study, we performed sequence analysis for the essential promoter region and the protein-binding pocket domain and observed RB1 missense mutations in five out of 27 (18.5%) dedifferentiated components, whereas no mutation was detected in the corresponding well-differentiated components. All the cases with RB1 mutation were LOH-positive and showed decreased pRB expression. The mutation rate of dedifferentiated component was almost equivalent to that of malignant fibrous histiocytoma reported by Chibon et al, but our control malignant fibrous histiocytoma cases showed no mutation whatsoever. This was perhaps because our malignant fibrous histiocytoma cases did not include cases which occurred in the retroperitoneum, or may simply have been because the number of our malignant fibrous histiocytoma cases was relatively small.
The RB1 gene harbors a small (almost 600 bp) CpG island that encompasses the essential promoter region. Experimental data have shown that in vitro methylation of the RB1 promoter region reduced pRB expression.28 Unilateral retinoblastoma and some brain tumors show loss of pRB expression which is associated with aberrant methylation of the CpG island within the RB1 promoter region;29, 30, 31 however, the methylation status of the RB1 gene in soft-tissue sarcoma has not been described. RB1 promoter hypermethylation was detected in four out of 27 (14.8%) dedifferentiated components, whereas no hypermethylation was detected in any of the well-differentiated components. In contrast to the results of mutation analysis, three out of the four cases with hypermethylation had no LOH.
pRB, encoded by the RB1 gene, is a key regulator of proliferation, development, and differentiation of certain cell types. pRB is phosphorylated and dephosphorylated synchronously with the cell cycle. The phosphorylation of pRB by the cyclin D1/CDK4(CDK6) complex in the late G1 phase results in the release of nuclear proteins and transcription factors, including the E2F family, thereby initiating the expression of genes critical for transition into the S phase of the cell cycle. It has been proved that MDM2 interacts physically and functionally with pRB and, as with p53 protein, inhibits the pRB regulatory function.32 Using immunohistochemical methods, the present study demonstrated the expression of pRb to be abnormal in 18 dedifferentiated components and in nine well-differentiated components, results which are similar to those observed in a previous study.7 These results suggest that pRB plays a role in dedifferentiation from well-differentiated liposarcoma to dedifferentiated liposarcoma, through the alteration of the coding gene RB1, or through the interaction of CDK4 or MDM2, which are considered to be expressed at a higher level in dedifferentiated component than in well-differentiated component, as described above. pRB has been shown to promote adipocyte differentiation by enhancing the DNA-binding and transactivation activity of C/EBPβ, and/or by inhibiting Ras signaling to suppress the activation of the ERK1/2 MAPKs.33, 34 These findings support our hypothesis.
In summary, we investigated the genetic alterations of the RB1 gene, in addition to the LOH status of 13q12–q14 and pRB expression, in two morphologically distinct areas (dedifferentiated component and well-differentiated component) in 27 dedifferentiated liposarcoma patients. LOH and abnormal pRB expression were observed more frequently in the dedifferentiated component than in the well-differentiated component. Five and four out of the 27 dedifferentiated component samples harbored mutations and promoter methylation, respectively, whereas the well-differentiated components showed no such alterations. These results suggest that pRB plays a role in ‘dedifferentiation’, and that ‘two-hit’ mechanism is involved in the altered pRB expression in dedifferentiated liposarcoma.
References
Henricks WH, Chu YC, Goldblum JR, et al. Dedifferentiated liposarcoma: a clinicopathological analysis of 155 cases with a proposal for an expanded definition of dedifferentiation. Am J Surg Pathol 1997;21:271–281.
Hasegawa T, Seki K, Hasegawa F, et al. Dedifferentiated liposarcoma of retroperitoneum and mesentery: varied growth patterns and histological grades—a clinicopathologic study of 32 cases. Hum Pathol 2000;31:717–727.
Pilotti S, Della Torre G, Lavarino C, et al. Distinct mdm2/p53 expression patterns in liposarcoma subgroups: implications for different pathogenetic mechanisms. J Pathol 1997;181:14–24.
Dei Tos AP, Doglioni C, Piccinin S, et al. Molecular abnormalities of the p53 pathway in dedifferentiated liposarcoma. J Pathol 1997;181:8–13.
Sakamoto A, Oda Y, Adachi T, et al. H-ras oncogene mutation in dedifferentiated liposarcoma. Polymerase chain reaction-restriction fragment length polymorphism analysis. Am J Clin Pathol 2001;115:235–242.
Sakamoto A, Oda Y, Adachi T, et al. Beta-catenin accumulation and gene mutation in exon 3 in dedifferentiated liposarcoma and malignant fibrous histiocytoma. Arch Pathol Lab Med 2002;126:1071–1078.
Schneider-Stock R, Boltze C, Jaeger V, et al. Significance of loss of heterozygosity of the RB1 gene during tumour progression in well-differentiated liposarcomas. J Pathol 2002;197:654–660.
Zheng L, Lee WH . The retinoblastoma gene: a prototypic and multifunctional tumor suppressor. Exp Cell Res 2001;264:2–18.
Cance WG, Brennan MF, Dudas ME, et al. Altered expression of the retinoblastoma gene product in human sarcomas. N Engl J Med 1990;323:1457–1462.
Wunder JS, Czitrom AA, Kandel R, et al. Analysis of alterations in the retinoblastoma gene and tumor grade in bone and soft-tissue sarcomas. J Natl Cancer Inst 1991;83:194–200.
Karpeh MS, Brennan MF, Cance WG, et al. Altered patterns of retinoblastoma gene product expression in adult soft-tissue sarcomas. Br J Cancer 1995;72:986–991.
Cohen JA, Geradts J . Loss of RB and MTS1/CDKN2 (p16) expression in human sarcomas. Hum Pathol 1997;28:893–898.
Wadayama B, Toguchida J, Shimizu T, et al. Mutation spectrum of the retinoblastoma gene in osteosarcomas. Cancer Res 1994;54:3042–3048.
Chibon F, Mairal A, Freneaux P, et al. The RB1 gene is the target of chromosome 13 deletions in malignant fibrous histiocytoma. Cancer Res 2000;60:6339–6345.
Chibon F, Mariani O, Derre J, et al. A subgroup of malignant fibrous histiocytomas is associated with genetic changes similar to those of well-differentiated liposarcomas. Cancer Genet Cytogenet 2002;139:24–29.
Coindre JM, Mariani O, Chibon F, et al. Most malignant fibrous histiocytomas developed in the retroperitoneum are dedifferentiated liposarcomas: a review of 25 cases initially diagnosed as malignant fibrous histiocytoma. Mod Pathol 2003;16:256–262.
Takahira T, Oda Y, Tamiya S, et al. Microsatellite instability and p53 mutation associated with tumor progression in dermatofibrosarcoma protuberans. Hum Pathol 2004;35:240–245.
Saito T, Oda Y, Kawaguchi K, et al. Possible association between tumor-suppressor gene mutations and hMSH2/hMLH1 inactivation in alveolar soft part sarcoma. Hum Pathol 2003;34:841–849.
Herman JG, Graff JR, Myohanen S, et al. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA 1996;93:9821–9826.
Karpeh MS, Brennan MF, Cance WG, et al. Altered patterns of retinoblastoma gene product expression in adult soft-tissue sarcomas. Br J Cancer 1995;72:986–991.
Weiss SW, Rao VK . Well-differentiated liposarcoma (atypical lipoma) of deep soft tissue of the extremities, retroperitoneum, and miscellaneous sites. A follow-up study of 92 cases with analysis of the incidence of ‘dedifferentiation’. Am J Surg Pathol 1992;16:1051–1058.
McCormick D, Mentzel T, Beham A, et al. Dedifferentiated liposarcoma. Clinicopathologic analysis of 32 cases suggesting a better prognostic subgroup among pleomorphic sarcomas. Am J Surg Pathol 1994;18:1213–1223.
Dal Cin P, Kools P, Sciot R, et al. Cytogenetic and fluorescence in situ hybridization investigation of ring chromosomes characterizing a specific pathologic subgroup of adipose tissue tumors. Cancer Genet Cytogenet 1993;68:85–90.
Rosai J, Akerman M, Dal Cin P, et al. Combined morphologic and karyotypic study of 59 atypical lipomatous tumors. Evaluation of their relationship and differential diagnosis with other adipose tissue tumors (a report of the CHAMP Study Group). Am J Surg Pathol 1996;20:1182–1189.
Fritz B, Schubert F, Wrobel G, et al. Microarray-based copy number and expression profiling in dedifferentiated and pleomorphic liposarcoma. Cancer Res 2002;62:2993–2998.
Rieker RJ, Joos S, Bartsch C, et al. Distinct chromosomal imbalances in pleomorphic and in high-grade dedifferentiated liposarcomas. Int J Cancer 2002;99:68–73.
Hostein I, Pelmus M, Aurias A, et al. Evaluation of MDM2 and CDK4 amplification by real-time PCR on paraffin wax-embedded material: a potential tool for the diagnosis of atypical lipomatous tumours/well-differentiated liposarcomas. J Pathol 2004;202:95–102.
Ohtani-Fujita N, Fujita T, Aoike A, et al. CpG methylation inactivates the promoter activity of the human retinoblastoma tumor-suppressor gene. Oncogene 1993;8:1063–1067.
Stirzaker C, Millar DS, Paul CL, et al. Extensive DNA methylation spanning the Rb promoter in retinoblastoma tumors. Cancer Res 1997;57:2229–2237.
Nakamura M, Yonekawa Y, Kleihues P, et al. Promoter hypermethylation of the RB1 gene in glioblastomas. Lab Invest 2001;81:77–82.
Gonzalez-Gomez P, Bello MJ, Alonso ME, et al. CpG island methylation status and mutation analysis of the RB1 gene essential promoter region and protein-binding pocket domain in nervous system tumours. Br J Cancer 2003;88:109–114.
Xiao ZX, Chen J, Levine AJ, et al. Interaction between the retinoblastoma protein and the oncoprotein MDM2. Nature 1995;375:694–698.
Chen PL, Riley DJ, Chen Y, et al. Retinoblastoma protein positively regulates terminal adipocyte differentiation through direct interaction with C/EBPs. Genes Dev 1996;10:2794–2804.
Hansen JB, Petersen RK, Jorgensen C, et al. Deregulated MAPK activity prevents adipocyte differentiation of fibroblasts lacking the retinoblastoma protein. J Biol Chem 2002;277:26335–26339.
Acknowledgements
The English used in this manuscript was revised by Miss K Miller (Royal English Language Centre, Fukuoka, Japan). This work was supported in part by a Grant-in-Aid for Scientific Research (C) (15590304) from the Japan Society for the Promotion of Science.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Takahira, T., Oda, Y., Tamiya, S. et al. Alterations of the RB1 gene in dedifferentiated liposarcoma. Mod Pathol 18, 1461–1470 (2005). https://doi.org/10.1038/modpathol.3800447
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.3800447
Keywords
This article is cited by
-
Molecular and clinicopathological analysis revealed an immuno-checkpoint inhibitor as a potential therapeutic target in a subset of high-grade myxofibrosarcoma
Virchows Archiv (2022)
-
Liposarcoma: a soft tissue tumor with many presentations
MUSCULOSKELETAL SURGERY (2014)
-
An experimental model for the study of well-differentiated and dedifferentiated liposarcoma; deregulation of targetable tyrosine kinase receptors
Laboratory Investigation (2011)
-
Low-grade dedifferentiated liposarcoma of the neck: magnetic resonance imaging and pathological correlation
Journal of Orthopaedic Science (2010)
-
Genomic profiling reveals subsets of dedifferentiated liposarcoma to follow separate molecular pathways
Virchows Archiv (2010)