Abstract
Serrated adenoma is a recently described entity characterized by having combined architectural features of hyperplastic polyps and classical adenoma. To understand the role of gene regulation in the progression of the serrated neoplasia pathway, we examined the methylation profiles of the promoter regions of 19 genes, DNA ploidy, and mutator phenotype status. In all, 40 sporadic, classical serrated adenomas were pathologically reviewed and divided into four pathologic groups according to their histologic grades. Methylation-specific PCR was performed using primers for p16, hMLH1, RASSF1A, APC, HIC-1, DAPK, MGMT, SLC5A8, RB1, H-Cadherin, E-Cadherin, TIMP3, PTEN, THBS1, LKB1, p14, p15, FHIT, and VHL. Dual flow-cytometric analyses using cytokeratin and DAPI and MSI studies using BAT26 were also performed. Methylation was observed in 2.5–82.5% (mean 33.9%) of the CpG islands in the promoter regions of 16 genes. The tumors with higher histologic grades, including carcinomas, showed more extensive methylation compared to those with lower grades, and serrated adenomas in the right colon showed more frequent methylation than those in the left (P<0.05). Tumor-specific promoter methylation of SLC5A8 was observed in 33 (82.5%) of the serrated adenomas. Aneuploidization with near-diploid DNA indices was detected in four out of 28 cases examined (14.3%); two were low-grade serrated adenomas and two were carcinomas in the left colon. The high mutator phenotype was not observed in any of the cases examined. Our results indicate that: (1) aberrant, widespread methylation of CpG islands increases with the histological progression of serrated adenomas; (2) methylation of SLC5A8 is an early event; and (3) additional methylation of the p16, p14, MGMT, TIMP3, and FHIT genes are important tumorigenic steps in the serrated neoplasia pathway.
Similar content being viewed by others
Main
Serrated adenoma is a recently defined colorectal neoplasm with a prevalence ranging from 0.6 to 1.3%.1, 2 The serrated neoplasia pathway suggests that dysplasia can arise within hyperplastic colonic polyps, resulting in the formation of a serrated adenoma and potentially the development of a colorectal carcinoma.2, 3, 4, 5 The process, presumably driven by a progressive accumulation of genetic changes within the clonal cell mass, is phenotypically characterized by increasing dysplasia in an epithelium that retains a distinctive serrated architecture.6 The polyps of this pathway differ morphologically and genetically from those of the traditional adenoma–carcinoma sequence.7 Genetically, serrated adenomas are differentiated from sporadic tubular adenomas by a high frequency of the low mutator phenotype,3, 8 p53 mutations,9 KRAS mutations,9, 10 BRAF mutations,11 and a low frequency of APC gene mutations.12 However, the prevalence of the high mutator phenotype and its significance during tumorigenesis are controversial and the chromosomal status of serrated adenoma has not been studied.
Methylation of CpG islands is a mechanism for the suppression of gene transcription in physiological and pathological settings, including neoplasia.13 The recently described CpG island methylator phenotype is an alternative tumorigenesis pathway characterized by methylation of multiple promoter regions of tumor suppressor genes harboring CpG islands. Recent studies have shown that CpG island methylator phenotype is very common in sporadic serrated adenomas, and especially in the hyperplastic polyps of hyperplastic polyposis.14, 15 However, the methylation profiles of tumor-related genes in the serrated neoplasia pathway during the progression of serrated adenomas remains to be studied.
In the present study, serrated adenomas selected according to strict criteria were histologically divided into low and high grades with or without carcinomas and their methylation status was studied using primers related to tumor suppressor genes, genes related to DNA repair or carcinogen detoxification, cell cycle regulators, an angiogenesis inhibitor, and invasion or metastasis suppressors. The methylation status of these genes was compared with the chromosomal aberration status and high mutator phenotype of the tumors.
Materials and methods
Materials
In all, 40 sporadic serrated adenomas were collected by polypectomy from patients at Our Lady of Mercy Hospital, The Catholic University of Korea, in Inchon, and Samsung Medical Center, Sungkyunkwan University, Seoul, from 1998 to 2003. The 40 patients included 14 females and 26 males with a mean age of 54.7 (range 35–86). In the present study, strict and objective histologic criteria were used for the diagnosis of serrated adenoma: serration in ≥20% of the lesions, frequent pseudostratification, nuclear atypia, and micropapillations of the surface epithelium with eosinophilic cytoplasm.7 All of our cases were the traditional type of serrated adenomas according to the provisional classification of serrated polyps by Torlakovic et al.16 The serrated adenomas were divided into four pathologic groups according to the presence of hyperplastic polyp components described by Longacre and Fenoglio-Preiser1 and epithelial atypia based on nuclear atypia (nuclear enlargement, hyperchromasia, prominent nucleoli, and presence of vesicular nuclei) and archictectural atypia (cribriform pattern and partial loss of polarity). The four groups were serrated adenomas with low histologic grade and hyperplastic area (N=5), serrated adenomas with low-grade atypism (N=23), serrated adenomas with high-grade atypism (N=6), and serrated adenomas with carcinomas (N=6). When there was low- and high-grade atypism within one polyp, it was regarded as high grade. Tumors larger than 0.5 cm were selected to ensure a sufficient amount of DNA for analysis. In 15 cases, normal colon tissue was obtained from the histologically normal portion of the polypectomy specimens (seven cases) or from patients without tumors (eight cases).
Methylation-Specific PCR
Methylation-specific PCR was performed using the primers described in Table 1. Briefly, DNA was extracted from 10 μm sections stained with 0.1% methylene blue or hematoxylin & eosin by manual microdissection under microscopy using 20-gauge needles and proteinase K solution as previously described.17 More than 40 000 cells were procured from each sample and more than 80% of these were tumor cells. After a bisulfite modification, β-actin was analyzed to standardize the DNA amount of each sample. PCR reactions were performed in a 25 μl reaction volume with 1.25 U of ExTaq polymerase (Takara Shuzo Co., Kyoto, Japan) as previously described.18 As a positive control, Sss-I methylase (New England Biolabs Inc, Beverly, MA, USA) was used to methylate 100 μg of DNA. The PCR products were electrophoresed in a 6% polyacrylamide gel, stained with SYBR Gold (Molecular Probes, Eugene, OR, USA), and then visualized under UV illumination.
The promoters of 19 genes were examined for their methylation status. These genes included tumor suppressor genes (p15INK4b, p16INK4a, HIC-1, APC, SLC5A8, PTEN, RB, FHIT, RASSF1A, VHL, LKB1), genes involved in DNA repair and the detoxification of carcinogens (hMLH1, O6-methylguanine-DNA-methyltransferase (MGMT)), cell cycle genes (PTEN, p14ARF), an angiogenesis inhibitor (TIMP3), and invasion or metastasis suppressors (E-cadherin, TIMP3, H-cadherin).
Flow Cytometry, Analysis of MSI, and Immunohistochemistry
Dual flow cytometric analyses using anti-cytokeratin (CAM5.2, Becton-Dickinson, CA, USA) and DAPI were performed as previously described.19 The ploidy level of cytokeratin-positive tumor cells was compared with benign, cytokeratin-negative diploid cells to create a DNA index. To allow for instrument error of 5%, the diploid tumors have DNA index of 0.95–1.05. All nondiploid tumors were analyzed twice.
To examine the microsatellite instability (MSI) status, we examined the tumor samples using the BAT26 primer set, which is a very sensitive marker of tumors with high MSI.20, 21 PCR analyses were performed using a DNA autosequencer (Applied Biosystems 373A sequencer; Applied Biosystems, CA, USA). The mobility shift of PCR products from the tumor DNA was compared to that of corresponding normal colonic mucosa. Immunohistochemistry was carried out on 4 μm paraffin sections with antibodies against hMLH1 (clone G168-15; 1:20 dilution) from BD Pharmingen (San Diego, CA, USA), and MGMT (clone MT3.1;1:50 dilution) and p16INK4a (p16INK4a Research Kit clone E6H4;1:30 Dilution) from DakoCytomation (Denmark). Sections were deparaffinized in xylene and rehydrated in graded series of alcohol to distilled water. Subsequently, antigen retreival by heating microwave oven in 10 mM citrate buffer (pH 6.0) was performed for 10 min. The detection was performed using a streptavidin-biotinylated horseradish peroxidase complex detection system (DakoCytomation, Denmark) according to the manufacturer’s instructions and visualized with 3,3′-diaminobenzidine as a substrate. The sections were counterstained with Mayer's hematoxylin.
Statistics
Statistical comparisons of the clinical, histologic, flow cytometry, mutator phenotype, immunohistochemistry, and methylation results were performed with the χ2 test and ANOVA using the statistical software SPSS (version 10.0, Chicago, IL, USA).
Results
Clinicopathologic Findings, MSI and Ploidy Status
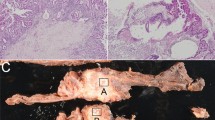
The serrated adenomas in this study met strict pathologic criteria and were polypoid and nodular in shape, with sizes ranging from 0.5 to 2.5 cm (mean 1.1 cm). Six cases occurred in the right colon and 34 occurred in the left colon. Of these 40 serrated adenomas, a low histologic grade of atypia was found in 28 cases, with five showing a hyperplastic area in the neck portion of the polyp (Figure 1). Representative examples of low- and high-grade atypism are depicted in Figure 2. Carcinomas arising from serrated adenomas were found in six cases (12.5%), including five intramucosal carcinomas and one deeply invasive carcinoma showing an invasion into pericolic adipose tissue by well-preserved serrated glands with a mucin pool (Figure 3). All carcinomas in the serrated adenomas were observed in the head portion of the polyp. In four of the 28 analyzed cases (14.3%), aneuploidization was detected using very sensitive, dual flow cytometry (Figure 4). All aneuploid tumors showed near-diploid DNA indices ranging 1.1–1.2. The four near-diploid tumors included two low-grade serrated adenomas and two carcinomas within serrated adenomas in the left colon. The high mutator phenotype was not observed in any of the cases (Figure 5).
Near-diploid tumor cells evaluated with dual flow cytometry using DAPI and cytokeratin. Upper left: total cell counts, Upper right: gating of cytokeratin-negative nontumorous cells, Lower right: gating of cytokeratin-positive tumor cells, Lower left: overlay of cytokeratin-positive and -negative cells.
Methylation Analyses of 19 Tumor-Related Genes
Methylation within serrated adenomas was observed in 16 promoter regions ranging from 2.5 to 82.5%. The overall methylation status are described in Table 2. In the normal colonic mucosa, methylation was observed in 13 promoter regions and ranged from 6.6 to 26.6%. Within the 15 samples of normal colonic mucosa, methylation was found with the indicated frequency in the promoters of p15 (4), SLC5A8 (2), H-cadherin (2), E-cadherin (2), TIMP3 (2), FHIT (2), DAPK (2), p14 (2), THBS (2), p16 (1), RASSF1A (1), APC (1), MGMT (1), RB1 (0), LKB1 (0), hMLH1 (0), HIC-1 (0), PTEN (0), and VHL (0). All promoter regions related to tumor suppressors were methylated significantly more in the tumors than in normal tissues.
No methylation was observed in the hMLH1, RB1, and VHL promoter regions from either the tumors or normal tissues. When the average number of genes methylated (percent methylation) was compared in each pathologic group, a stepwise increase was observed during the progression of the serrated neoplasia pathway (P<0.05) (Table 3). There were statistically significant differences in the methylation of serrated adenomas in the ‘low’ and ‘carcinoma’ groups and the ‘serrated adenomas’ ‘carcinoma’ groups (P<0.05). The methylation of the p16, MGMT, TIMP3, p14, and FHIT genes was also correlated with the progression of histologic grades (Table 3). Neither the percent methylation nor the methylation of individual genes was related to the patients’ gender, age, size of tumors, or ploidy status. The right colon showed a higher frequency of methylation for the p16, p14, TIMP3, and FHIT genes (P<0.05). Also, the overall percent methylation was higher in the right colon (36.8%) than the left (27.1%).
A high percentage of promoter methylation of SLC5A8 was detected in 33 serrated adenomas (82.5%) (Figure 6). Only two of the 15 (13.3%) normal colon tissue samples showed methylation of SLC5A8. A review of the colonic control samples with methylated SLC5A8 showed relatively hyperplastic crypts in the superficial portion of the mucosae. All four aneuploid tumors and two serrated adenomas in the right colons of female patients showed methylation of SLC5A8 gene. To evaluate the role of methylation in the pathogenesis of the serrated neoplasia pathway, 10 sporadic hyperplastic polyps were further evaluated for the presence of SLC5A8 methylation. Seven cases of sporadic hyperplastic polyps showed methylation of SLC5A8. p16 methylation was observed in nine of the 40 serrated adenomas (22.5%) including five with high-grade and four with low-grade atypism. Notably, methylation of p16 was found in seven cases with SLC5A8 methylation (Figure 6) and p16 methylation was significantly correlated with the methylation status of the HIC-1, TIMP3, p14, and FHIT genes (P<0.05). To evaluate the importance of methylation during tumorigenesis, immunohistochemistry was performed using available antibodies against hMLH1, MGMT, and p16. The promoter methylation status completely or partially matched the expression of these proteins: there was no loss of hMLH1 and partial losses of MGMT and p16.
Representative examples of methylation-specific PCR analysis of SLC5A8 (a) and p16 (b) for low-grade serrated adenomas (#32 and #34), a high-grade serrated adenoma (#36), and a carcinoma with serrated adenoma (#9). L: Marker, U: unmethylated, M: methylated, IVC: in vitro methylated positive control, N: negative control.
Discussion
In this study, we examined the promoter regions of 19 tumor-related genes. We found that the methylation of the promoter regions of p16, MGMT, TIMP3, p14, and FHIT genes is related to the histologic progression of serrated adenomas, and the methylation of the SLC5A8 gene occurs early in the serrated neoplasia pathway. SLC5A8 has recently been identified as a candidate tumor-suppressor gene that is involved in sodium transport. The silencing of SLC5A8 by methylation confers a specific growth advantage in the subset of colon cancers in which this locus is inactivated.22 It is methylated in 59% of primary colonic carcinomas and classical tubular adenomas but is rarely methylated in normal colonic mucosae. A high frequency of SLC5A8 methylation was identified in the serrated adenomas (82.5%) regardless of the histologic grade or clinicopathologic parameters. Hypermethylation of SLC5A8 was observed in all cases of serrated adenomas with hyperplastic areas and in 70% of hyperplastic polyps. In previous methylation studies using sporadic hyperplastic polyps, Chan et al15 did not detect any methylation within five promoter regions, including those of p16, MINT1, MINT2, MINT31, and hMLH1, while Bariol et al23 found methylation of these genes using fresh DNA from hyperplastic polyps. The tumor-specific methylation of SLC5A8 found in hyperplastic polyps and serrated adenomas with low-grade atypism suggests that methylation of CpG islands occurs early during the serrated neoplasia pathway. Although the promoters of 16 of the tumor-related genes were methylated, neither hMLH1 methylation nor a high mutator phenotype was found in this study. This low frequency of MSI may be due to the use of a single microsatellite marker, the predominantly left location of the cases used (85%), or the very low frequency of MSI-high colorectal carcinoma and hMLH1 methylation among elderly subjects in Korea.24 Our result is compatible with previous results reporting that high MSI and hMLH1 methylation are uncommon in serrated adenomas. Most high-MSI tumors are admixed hyperplastic and adenomatous polyps rather than serrated polyps.3, 4, 8, 11, 14, 15 In our study, the right colon showed a higher percentage of methylation and more frequent methylation of the p16, p14, TIMP3, and FHIT genes than the left colon. These results are comparable with the results reported by Burri et al25 using colonic carcinoma samples. It is believed that serrated adenomas and hyperplastic polyps occurring in the proximal colon in elderly females are at particular risk of progression via the serrated neoplasia pathway.26 Both of the female patients with serrated adenomas in the right colon showed a high percentage of methylation, suggesting a close relation between methylation and a higher possibility of malignant transformation in this situation.
To date, methylation studies on serrated adenomas have been limited due to the rare occurrence of serrated adenomas and the large amounts of DNA that are needed for methylation studies.14, 15 Serrated adenomas larger than 0.5 cm were selected in this study, with 21 larger than 1 cm in diameter. As a result, a large-scale methylation study has been achieved using these serrated adenomas.
Although Iwabuchi et al27 first reported aneuploidy in two serrated adenoma cases using an image analysis, DNA ploidy studies in serrated adenomas using flow cytometry have not been reported previously. We found that the frequencies of aneuploidization within serrated adenomas are similar to the results from conventional classical adenomas,28 as detected with a highly sensitive crypt isolation technique and cytometric analysis. However, the serrated adenomas exclusively showed near-diploid DNA indices rather than the high aneuploid indices reported in traditional tubular or villous adenomas. Genetic instability of tumors is proportional to their degree of aneuploidy,29, 30 so the near-diploid DNA indices observed in our four aneuploid serrated adenomas and carcinomas suggest that subtle chromosomal changes may underlie the serrated neoplasia pathway. Aneuploidy observed in serrated adenomas with both low-grade atypism and carcinomas also suggests that aneuploidization occurs early during tumor progression. When examining the results obtained from our methylation study and the flow cytometric analyses, it seems clear that the widespread hypermethylation of promoters and the subtle chromosomal aberrations are early events in the progression of the serrated neoplasia pathway.
Here, we have reported frequent, tumor-specific methylation of SLC5A8; methylation of the p16, MGMT, TIMP3, p14, and FHIT genes associated with histologic progression; and no hMLH1 methylation in serrated adenomas. The correlation of aberrant, progressive, widespread CpG-island methylation with the histologic progression and aneuploidization that occurs in low-grade serrated adenomas suggests these events occur early and are stable during the progression of the serrated neoplasia pathway.
References
Longacre TA, Fenoglio-Preiser CM . Mixed hyperplastic adenomatous polyps/serrated adenomas. A distinct form of colorectal neoplasia. Am J Surg Pathol 1990;14:524–537.
Jass JR . Serrated adenoma and colorectal cancer. J Pathol 1999;187:499–502.
Iino H, Jass JR, Simms LA, et al. DNA microsatellite instability in hyperplastic polyps, serrated adenomas, and mixed polyps: a mild mutator pathway for colorectal cancer? J Clin Pathol 1999;2:5–9.
Jass JR, Young J, Leggett BA . Hyperplastic polyps and DNA microsatellite unstable cancers of the colorectum. Histopathology 2000;37:295–301.
Jass JR . Serrated route to colorectal cancer: back street or super highway? J Pathol 2001;193:283–285.
Jass JR . Progress in gastrointestinal pathology in the genetic era. Pathology 2002;34:493.
Bariol C, Hawkins NJ, Turner JJ, et al. Histopathological and clinical evaluation of serrated adenomas of the colon and rectum. Mod Pathol 2003;16:417–423.
Jass JR, Biden KG, Cummings MC, et al. Characterization of a subtype of colorectal cancer combining features of the suppressor and mild mutator pathways. J Clin Pathol 1999;52:455–460.
Hiyama T, Yokozaki H, Shimamoto F, et al. Frequent p53 gene mutations in serrated adenomas of the colorectum. J Pathol 1998;186:131–139.
Higashidani Y, Tamura S, Morita, et al. Analysis of K-ras codon 12 mutation in flat and nodular variants of serrated adenoma in the colon. Dis Colon Rectum 2003;46:327–332.
Chan TL, Zhao W, Leung SY, et al. BRAF and KRAS mutations in colorectal hyperplastic polyps and serrated adenomas. Cancer Res 2003;63:4878–4881.
Dehari R . Infrequent APC mutations in serrated adenoma. Tohoku J Exp Med 2001;193:181–186.
Baylin SB, Herman JG, Graff JR, et al. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res 1998;72:141–196.
Park SJ, Rashid A, Lee JH, et al. Frequent CpG island methylation in serrated adenomas of the colorectum. Am J Pathol 2003;162:815–822.
Chan AO, Issa JP, Morris JS, et al. Concordant CpG island methylation in hyperplastic polyposis. Am J Pathol 2002;160:529–536.
Torlakovic E, Skovlund E, Snover DC, et al. Morphologic reappraisal of serrated colorectal polyps. Am J Surg Pathol 2003;27:65–81.
Kim KM, Salovaara R, Meckli JP, et al. PolyA deletions in hereditary nonpolyposis colorectal cancer: mutations before a gatekeeper. Am J Pathol 2002;160:1503–1506.
Dong SM, Kim HS, Rha SH, et al. Promoter hypermethylation of multiple genes in carcinoma of the uterine cervix. Clin Cancer Res 2001;7:1982–1986.
Glogovac JK, Porter PL, Banker DE, et al. Cytokeratin labeling of breast cancer cells extracted from paraffin-embedded tissue for bivariate flow cytometric analysis. Cytometry 1996;24:260–267.
Myeroff LL, Parsons R, Kim SJ, et al. A transforming growth factor beta receptor type II gene mutation common in colon and gastric but rare in endometrial cancers with microsatellite instability. Cancer Res 1995;55:5545–5547.
Halling KC, Harper J, Moskaluk CA, et al. Origin of microsatellite instability in gastric cancer. Am J Pathol 1999;155:205–211.
Li H, Myeroff L, Smiraglia D, et al. SLC5A8, a sodium transporter, is a tumor suppressor gene silenced by methylation in human colon aberrant crypt foci and cancers. Proc Natl Acad Sci USA 2003;100:8412–8417.
Bariol C, Suter C, Cheong K, et al. The relationship between hypomethylation and CpG island methylation in colorectal neoplasia. Am J Pathol 2003;162:1361–1371.
Kim HC, Kim CN, Yu CS, et al. Methylation of the hMLH1 and hMSH2 promoter in early-onset sporadic colorectal carcinomas with microsatellite instability. Int J Colorectal Dis 2003;18:196–202.
Burri N, Shaw P, Bouzourene H, et al. Methylation silencing and mutations of the p14ARF and p16INK4a genes in colon cancer. Lab Invest 2001;81:217–229.
Hawkins NJ, Bariol C, Ward RL . The serrated neoplasia pathway. Pathology 2002;4:548–555.
Iwabuchi M, Sasano H, Hiwatashi N, et al. Serrated adenoma: a clinicopathological, DNA ploidy, and immunohistochemical study. Anticancer Res 2000;20:1141–1147.
Sugai T, Takahashi H, Habano W, et al. Analysis of genetic alterations, classified according to their DNA ploidy pattern, in the progression of colorectal adenomas and early colorectal carcinomas. J Pathol 2003;200:168–176.
Duesberg P, Rausch C, Rasnick D, et al. Genetic instability of cancer cells is proportional to their degree of aneuploidy. Proc Natl Acad Sci USA 1998;95:13692–13697.
Lengauer C, Kinzler KW, Vogelstein B . Genetic instabilities in human cancers. Nature 1998;396:643–649.
Acknowledgements
This work was supported by research Grants 0210050 from the National Cancer Center of Korea to Dong SM and The Catholic University of Korea to Kim KM.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Dong, S., Lee, E., Jeon, E. et al. Progressive methylation during the serrated neoplasia pathway of the colorectum. Mod Pathol 18, 170–178 (2005). https://doi.org/10.1038/modpathol.3800261
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.3800261
Keywords
This article is cited by
-
Correlation between MGMT promoter methylation and response to temozolomide-based therapy in neuroendocrine neoplasms: an observational retrospective multicenter study
Endocrine (2018)
-
Distinct histopathological characteristics in colorectal submucosal invasive carcinoma arising in sessile serrated adenoma/polyp and conventional tubular adenoma
Virchows Archiv (2018)
-
O6-Methylguanine-DNA methyltransferase status in neuroendocrine tumours: prognostic relevance and association with response to alkylating agents
British Journal of Cancer (2015)
-
DNA methylation alterations of AXIN2 in serrated adenomas and colon carcinomas with microsatellite instability
BMC Cancer (2014)
-
A significant imbalance in mitosis versus apoptosis accelerates the growth rate of sessile serrated adenoma/polyps
Virchows Archiv (2013)