Abstract
Ki67 is a nuclear protein that is tightly linked to the cell cycle. It is a marker of cell proliferation and has been used to stratify good and poor prognostic categories in invasive breast cancer. Its correlation with gene expression patterns has not been fully elucidated. In this study, Ki67 immunohistochemistry using the MIB-1 antibody was performed on sections cut from 21 formalin-fixed, paraffin-embedded invasive breast cancers. Scoring was determined as nil (no immunostaining), low (10% or less immunopositivity) or high (>10% immunoreactive cells) respectively. The relationship of Ki67 immunohistochemical detection with clinicopathologic parameters was evaluated. Using Affymetrix U133A GeneChips, expression profiles for these tumors were generated and correlated with Ki67 immunohistochemical findings. Analysis of variance was used to define genes that were differentially regulated between the groups. Real-time polymerase chain reaction (PCR) was used to confirm the presence of a downregulated gene. Our results showed high, low and nil Ki67 immunostaining in nine (43%), six (28.5%) and six (28.5%) invasive breast cancers respectively, with increased Ki67 protein expression correlating with high histologic grade (P=0.02), mitotic score (P=0.001) and estrogen receptor immunonegativity (P=0.002). Expression profiling trends of the Ki67 gene mirrored the observed proportions of immunostained cells when the Ki67 immunoscore was >10%. Genes related to apoptosis and cell death (bcl2, MAP2K4, TNF10) were noted to be downregulated in tumors that disclosed >40% Ki67 immunostaining (P<0.001). Downregulation of the bcl2 gene was confirmed at the RNA level by real-time RT-PCR. Differential regulation of these genes, especially bcl2, may contribute to the biological nature of clinically more aggressive and highly proliferative breast cancers.
Similar content being viewed by others
Main
Ki67 is a labile, nonhistone nuclear protein that is tightly linked to the cell cycle. It is expressed in proliferating cells during mid G1 phase, increasing in level through S and G2, and peaking in the M phase of the cell cycle.1 It is rapidly catabolized at the end of the M phase, and is undetectable in resting (G0 and early G1) cells.2 Ki67 expression shows a good relationship with growth fraction in several model systems,3, 4 and does not appear to be expressed during DNA repair processes. Hence, it is regarded as a marker of cell proliferation, and in invasive breast cancer, has been used to stratify patients into good and poor prognostic categories.2 It has also been reported to correlate with clinical response to chemotherapy.5, 6
The appropriate cutoff values that distinguish between high and low proliferative activity in a clinically relevant manner using Ki67 immunohistochemistry in breast cancer, however, has not been universally established.7, 8 The relationship of Ki67 protein expression with gene expression profiles is also not fully investigated. In this study, we evaluate Ki67 immunohistochemical protein detection in a series of invasive breast cancers using the MIB-1 antibody that recognizes Ki67 in formalin-fixed, paraffin-embedded tissue,4 and compare the findings against expression profiles using high-density oligonucleotide microarrays.
Materials and methods
Patients and Tumors
Breast tissues were obtained from the National Cancer Centre (NCC) Tissue Repository, with approvals from both the NCC Tissue Repository and Ethics committees. There was informed patient consent. The breast specimens were macroscopically assessed immediately after surgery, and a portion of tumor was flash-frozen in liquid nitrogen. The tissues were then fixed in 10% buffered formalin and subjected to routine pathological examination. Tumor size, histological subtype and grade,9 lymph node stage and lymphovascular invasion were documented. Histological grading was based on a composite score derived from assessment of three parameters: mitotic activity, nuclear pleomorphism and the extent of tubule formation.9, 10 A mitotic count of 0–4, 5–9, ≥10 mitoses per 10 high power fields (hpfs) gave mitotic scores of 1, 2 and 3, respectively, calibrated from the diameter of the microscope high-power field.9 An aggregate score of 3–5 was regarded as grade 1, 6–7 as grade 2, and 8–9 as grade 3. Lymph node stage was defined as 1 for node-negative disease, stage 2 for up to three axillary nodes involved by metastases, and stage 3 for 4 or more positive nodes.11
Immunohistochemistry
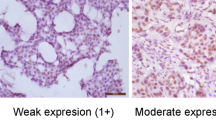
Sections of 4 μm thickness were cut from representative paraffin blocks containing invasive cancer, and immunohistochemistry for Ki67 (clone MIB-1, DAKO M7240, dilution 1:70) was carried out, with antigen retrieval using pressure cooking in 0.01 M citrate buffer at pH 6.0, and detection via DAKOCytomation envision/HRP kit K5007. Only distinct nuclear staining of invasive carcinoma cells was used for scoring via the light microscope, which was determined semiquantitatively as nil (no immunostaining), low (10% or less immunopositivity) or high (>10% immunoreactive cells) respectively.12 Assessment was carried out on the entire tumor represented in the section.
Immunohistochemistry for estrogen receptor (ER, Clone 6F11, Ventana 760-2596, prediluted), progesterone receptor (PR, Clone 1A6, Ventana 760-2547, prediluted) and cerbB2 (NeoMarker MS730-P, Clone e2-4001+3B5, 1:700 dilution) was also determined. For ER and PR, antigen retrieval was accomplished by pressure cooking in a microwave oven (using milestone T/T Mega) at 120°C for 4 min in citrate buffer at pH 6; while that for cerbB2 involved microwaving for 20 min in citrate buffer at pH 6. The proportion and intensity (0, 1+, 2+ and 3+) of immunostaining was evaluated. For ER and PR, ≥10% nuclear staining of at least 2+ intensity was required for a positive result.13 CerbB2 immunostaining was classified following the criteria recommended by the DAKO protocol.14
Appropriate positive and negative controls were run with each batch; benign breast epithelium within the sections served as internal positive controls for ER and PR.
Microarray Hybridization
RNA was extracted from frozen tumor tissue using TRIzol, and processed according to the manufacturer's instructions (Affymetrix). The extracted RNA was first passed through a Qiagen polymerase chain reaction (PCR) purification spin column, and the quality checked by OD258/280, which was >1.8.
Affymetrix chips were processed according to standard procedures. Raw chip scans were quality controlled using Genedata Refiner, and analyzed using Genedata Expressionist. With these Affymetrix U133A GeneChips, expression profiles for the tumors were generated and correlated with the Ki67 status as determined on immunohistochemistry.
Quality Controls and Normalization of Expression
After generating expression profiles of these tumors, the overall profile of each tumor was checked against one another to confirm that the range of expression values for each sample was comparable. These profiles were then internally normalized so that the median expression value of each profile was 750.
Real-Time Reverse Transcriptase (RT)-PCR
Appropriate primers were selected using the primer design software PRIMER3. The primer sequence for bcl2 was: forward primer: 5′-TCATGAAATATGCA TCTCACTGG-3′, reverse primer: 5′-AAATGCAATC CACTGTCACTCTT-3′. BLASTing of primers was carried out to confirm uniqueness. Reverse transcription using commercially available reverse transcriptase (Gibco-BRL) was carried out with 2 μg total RNA, 2 μl random hexamer (50 ng/μl), 1 μl oligo-dT15 (0.5 μg/μl), and topped up with DEPC H2O to 10 μl. The mixture was then heated to 70°C for 10 min, with subsequent cooling to room temperature. Optimization of PCR was carried out using different concentrations of MgCl2 and primers. PCR was performed using a Light Cycler (Roche) in a total reaction mixture containing Lightcycler-FastStart DNA Master SYBR Green I. After denaturation at 95°C for 10 min, 45 cycles were carried out at 95°C for 10 s, 58°C for 5 s, and 72°C for 8 s. A no template control was included in each run and 18 s rRNA control from genomic DNA isolated from human placenta was used for normalization of target genes. To verify the specificity of the amplification reaction, melting curve analysis was performed. Relative quantification was calculated using the comparative threshold (CT) method where ΔCT equals the difference between the CT values of the target gene and 18 s RNA. The CT value is taken as the fractional cycle number at which the emitted fluorescence of the sample passes a fixed threshold above the baseline. A series of samples corresponding to high and nil Ki67 immunostaining were selected for the real-time PCR assay.
Computational Analysis with STRING Database
The STRING database15 was used to predict if there were any functional associations of differentially regulated genes with Ki67 protein and gene expression. This database uses three predicting algorithms that are based on: (a) discovering gene fusion events;16, 17 (b) similar species coverage for functionally associated genes18 and (c) identification of genes that are close neighbors in a genome.19 STRING implements a scoring-framework based on benchmarks of the different types of associations. The database contains 444 238 genes in 110 species as of May 2004.
Statistical Analysis
The results were analyzed using the statistical software SPSS for Windows, version 11. The association of Ki-67 immunohistochemical detection with clinicopathologic parameters was studied using the χ2 test and frequency distributions were carried out using descriptive statistics. (Analysis of variance) ANOVA was used to define genes that were differentially regulated between the groups.
Results
The clinicopathologic parameters and Ki67 immunohistochemical results of the breast tumors are detailed in Table 1. All the women were of Chinese ethnicity except for one Malay (case 12). Their ages ranged from 40 to 79 years (mean and median 56 years). Mean and median tumor sizes were 38.5 and 40 mm, respectively. The tumor subtype was infiltrative ductal carcinoma in all but one (case 17), which was an invasive lobular carcinoma. Ki67 immunostaining scores revealed a mean of 22% and a median of 10% (Figure 1).
Correlation of Ki67 Immunostaining with Clinicopathologic Parameters
There was a statistically significant association of Ki67 immunohistochemical staining with mitotic score (P=0.001), histologic grade (P=0.02) and ER status (P=0.002). No correlation was found with age, tumor size, lymphovascular invasion, axillary nodal status, PR or cerbB2 status. As the patients were predominantly of Chinese ethnicity and tumors of infiltrative ductal subtype, statistical correlations were not pursued for these two variables.
Correlation of Gene Expression to Ki67 Immunohistochemistry
The Ki67 gene was identified in the tumor profiles and compared against the three categories of Ki67 protein expression as detected by immunohistochemistry: nil, low and high proliferative activity, respectively. These expression profiles are depicted in Figure 2. There was a correlation between Ki67 gene and protein expression. The Ki67 gene was most highly expressed in case 8, which demonstrated the highest percentage of Ki67 positive cells (90%) on immunohistochemistry. Among cases classified as showing high proliferative activity (>10% Ki67 immunostained cells), we observed a decrease in Ki67 gene expression as the percentage of Ki67 immunopositive cells diminished (P<0.05). This trend, however, was less evident in tumors immunohistochemically classified as disclosing low proliferative activity, while three cases without detectable Ki67 immunopositivity did not express the Ki67 gene.
To identify genes that were correlated to Ki67 protein expression, we defined two groups of tumors, one consisting of tumors containing ≥40% Ki67 positive cells (cases 1, 2, 3, 4, 8) and another group comprising tumors in which Ki67 immunohistochemistry was negative (cases 17, 18, 20). Based on the range of Ki67 protein expression values attained in the series, an arbitrary cutoff of 40% immunostained cells was used in order to include approximately half of the tumors categorized into the high proliferative activity group for identification of correlated genes.
ANOVA was utilized to define genes which were differentially regulated between these two groups at a confidence level of P<0.001 and which exhibited a mean-fold change of 2 between the groups.
A total of 28 genes were identified, of which five were upregulated in tumors demonstrating ≥40% Ki67 immunopositivity, while the remaining 23 genes were downregulated. These genes are listed in Table 2. Among the downregulated genes, many were related to cell death (bcl2, MAP2K4, TNFSF10).
Verification of bcl2 Messenger RNA Downregulation by Real-Time PCR
This was used to confirm one of the downregulated genes, bcl2 (Figures 3 and 4), with the real-time data correlating very well with the Affymetrix results. Table 3 shows the relative quantification of bcl2 and 18S rRNA on three cases classified as high proliferative activity, and two cases without Ki67 immunostaining, respectively.
Functional Associations of bcl2 in Tumors with Upregulation of the Ki67 Gene by Computational Analysis
Using computational analysis for the three cell-death-associated downregulated genes, those genes predicted to have functional associations with bcl2 in highly proliferative tumors are shown in Table 4, with scores derived from ‘text mining’. STRING did not find evidence of interaction from either ‘neighborhood’, ‘fusion’ or ‘occurrence’ methods. No significant functional predictions were found for MAP2K4 and TNFSF10.
Discussion
Routine assessment of cell proliferation is recommended in the pathologic evaluation for all breast cancers.2 This has traditionally taken the form of mitotic activity scoring, which is an integral component of histologic grading, an established prognostic marker in breast cancer.20 While the role of Ki67 immunohistochemistry as a prognostic and predictive marker in breast cancer has been widely investigated,5, 6, 8, 21, 22, 23, 24, 25 staining thresholds and cutoffs have been mostly arbitrary and are not universally standardized. Percent positively stained nuclei,26, 27 number of nuclei stained per 10 high power fields,5 stratification of MIB-1 staining in tertiles8 have been described. Some authors use Ki67 staining of 20% or more to define a high level,6 while others advocate segregating high from low staining via the median of positively stained cells as the distinguishing value.25 Recently, Spyratos et al7 have suggested that the optimal threshold level should be 25% based on multivariate analyses of MIB-1 immunostaining at different cutoffs. Additional variation issues include selection of the appropriate area for evaluation of immunostaining, be it the center or periphery of the tumor; area of highest cell density; or zone of highest tumor cell immunohistochemical reactivity.2
In this study, we used a scoring system described previously in our experience with Ki67 immunohistochemistry in breast ductal carcinoma in situ.12 Interestingly, the cutoff of 10% as the threshold between high and low proliferative activity coincided with the median value of immunostaining in this current series. With this scheme, we found a statistically significant association of Ki67 protein expression with mitotic score, histologic grade and negative ER status. The correlation with mitotic score and histologic grade is expected, as both Ki67 and mitotic activity are parallel markers of cell proliferation, and histologic grade has mitotic score as one of its component elements. The inverse relationship of high Ki67 immunostaining with negative ER immunohistochemistry is also consonant with higher grade, highly proliferative tumors being less likely to express ER; and our previous study of invasive breast cancers in our local population had confirmed a statistically significant inverse association between histological grade and ER status.13
Among the cases that demonstrated high proliferative activity based on Ki67 immunostaining of >10%, case 5 showed a very low mitotic score of only 1 mitosis per 10 hpfs, despite a Ki67 immunoscore of 30%. That the Ki67 immunostaining was a true reflection of protein expression may be partly substantiated by a corresponding increase in gene expression. One possible reason for this discordance between mitotic score vs Ki67 protein and gene expression is fixation delay, which is known to lead to loss of mitotic figures on histology.28 Also, the mitotic phase represents the smallest portion of cycling cells, and Ki67 immunohistochemistry, which detects cells from mid-G1 through S and G2 phases, will also decorate cells that do not manifest mitotic figures.
While our somewhat arbitrary scheme is validated by the association with morphologic and biological (ER) parameters, the correlation with Ki67 gene expression also lends weight to the basis for this scoring method. Nevertheless, the trend for a direct correlation between gene and protein expression appears to be more robust only for cases that were scored as displaying >10% Ki67 immunopositivity.
When Ki67 immunohistochemical staining results were compared against other genes that were differentially regulated, it was found that at a cutoff of 40% Ki67 immunopositivity, intriguingly, a number of genes related to cell death (bcl2, MAP2K4, TNFSF10) were downregulated. The differential regulation of these genes may contribute to the biological nature of the high Ki67 tumors, which are regarded as clinically more aggressive.2, 8
As bcl2 is a well-established gene that has been reported to be of prognostic import in breast cancer,2 we decided to study its association with Ki67 expression further by verifying its downregulation by real-time RT-PCR, which showed good correlation with the expression profiles. The bcl2 proto-oncogene was first described in follicular B-cell lines as a result of a translocation that juxtaposed the bcl2 gene on chromosome 18 with the immunoglobulin heavy-chain (IgH) gene on chromosome 14, t(14;18); leading to bcl2 protein overexpression due to deregulation of the bcl2–IgH fusion gene, allowing lymphocytes to become resistant to apoptosis or programmed cell death.29 However, bcl2 is also known to have an inhibitory effect on the cell cycle.30, 31, 32 It has been suggested that bcl2 represses growth by either lengthening the G1 phase30 or the rate at which cells transit from G0 into S phase.31, 33 It is also plausible that the growth inhibitory function of bcl2 is mediated by its functional association with molecules predicted by the STRING database such as cyclin D1, which is known to cause growth suppression.34 It has been previously demonstrated that besides enhancing mitogen-independent cell proliferation, c-myc, another gene that is functionally related to bcl2 according to the STRING database, can also suppress bcl2 expression.35, 36 In light of the above, a decrease in bcl2 would promote cell proliferation, which is in accord with our finding that this oncogene is downregulated in highly proliferative breast tumors. Conversely, bcl2 overexpression would therefore be a marker of improved prognosis in breast cancer, a finding that has been confirmed by other investigators.37
MAP2K4 is a mitogen-activated protein kinase kinase (MAPKK) homolog, which is a mediator of the stress-activated protein kinase (SAPK) pathways.38 However the SAPK pathways' role as a tumor-suppressive system, although promising, is still not well defined. The TNFSF10 gene that encodes tumor necrosis factor (ligand) superfamily, member 10, and also known as tumor necrosis factor-related apoptosis-inducing ligand (TRAIL), has been shown to suppress the outgrowth of human prostate tumor xenografts in SCID mice.39 By binding selectively to death receptors in cancer cells, the cytotoxic activity of TRAIL holds promise as an anticancer agent.40 It is interesting that TNFSF10 is downregulated in highly proliferative breast cancers and more work should be done to explore this new finding.
In conclusion, our study has shown that immunohistochemical protein detection of Ki67 in invasive breast cancer correlates with Ki67 gene expression profiles only when immunohistochemical reactivity is >10% of tumor cell nuclei stained, validating at the molecular level, the usage of this cutoff in immunohistochemical scoring systems for Ki67. However, the association with downregulation of bcl2, MAP2K4 and TNFSF10 is observed only beyond a threshold of 40%, suggesting that a certain level of proliferative activity has to be achieved in order to activate the trigger for downregulation of genes related to apoptosis and cell death. Further studies to investigate this relationship between cell proliferation as defined by Ki67 expression, and such apoptotic and cell death-related genes, will enhance understanding of the pathogenesis of breast cancer, and lend insights into potential therapeutic modulations.
References
Gerdes J, Lemke H, Baisch H, et al. Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. J Immunol 1984;133:1710–1715.
Fitzgibbons PL, Page DL, Weaver D, et al. Prognostic factors in breast cancer. College of American Pathologists Consensus Statement 1999. Arch Pathol Lab Med 2000;124:966–978.
Scott RJ, Hall PA, Haldane JS, et al. A comparison of immunohistochemical markers of cell proliferation with experimentally determined growth fraction. J Pathol 1991;165:173–178.
McCormick D, Chong H, Hobbs C, et al. Detection of the Ki-67 antigen in fixed and wax embedded sections with the monoclonal antibody Mib1. Histopathology 1993;22:355–360.
Archer CD, Parton M, Smith IE, et al. Early changes in apoptosis and proliferation following primary chemotherapy for breast cancer. Br J Cancer 2003;89: 1035–1041.
Petit T, Wilt M, Velten M, et al. Comparative value of tumor grade, hormonal receptors, Ki-67, HER-2 and topoisomerase II alpha status as predictive markers in breast cancer patients treated with neoadjuvant anthracycline-based chemotherapy. Eur J Cancer 2004;40: 205–211.
Spyratos F, Ferrero-Pous M, Trassard M, et al. Correlation between MIB-1 and other proliferation markers: clinical implications of the MIB-1 cutoff value. Cancer 2002;94:2151–2159.
Offersen BV, Sorensen FB, Knoop A, et al. Danish Breast Cancer Cooperative Tumor Biology Committee. The prognostic relevance of estimates of proliferative activity in early breast cancer. Histopathology 2003;42: 573–582.
Sloane JP, Anderson TJ, Blamey RW, et al. Pathology Reporting in Breast Cancer Screening, 2nd edn. National Coordinating Group for Breast Screening Pathology 1995, NHSBSP Publication No. 3, Sheffield, pp 19–21.
Elston CW . Grading of invasive carcinoma of the breast In: Page DL, Anderson TJ (eds). Diagnostic Histopathology of the Breast. Churchill Livingstone: New York, 1987, pp 300–311.
Pinder SE, Ellis IO, Elston CW . Prognostic factors in invasive breast cancer. J Cell Pathol 1998;3:57–62.
Tan PH, Chuah KL, Chiang G, et al. Correlation of p53 and cerbB2 expression and hormonal receptor status with clinicopathologic parameters in ductal carcinoma in situ of the breast. Oncol Rep 2002;9:1081–1086.
Thike AA, Chng MJ, Fook-Chong S, et al. Immunohistochemical expression of hormone receptors in invasive breast cancer: correlation of results of H-score with pathologic parameters. Pathology 2001;33:21–25.
Selvarajan S, Bay BH, Choo A, et al. Effect of fixation period on HER2/neu gene amplification detected by fluorescence in situ hybridization in invasive breast carcinoma. J Histochem Cytochem 2002;50:1693–1696.
von Mering C, Huynen M, Jaeggi D, et al. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res 2003;31:258–261.
Enright AJ, Iliopoulos I, Kyrpides NC, et al. Protein interaction maps for complete genomes based on gene fusion events. Nature 1999;402:86–90.
Marcotte EM, Pellegrini M, Ng HL, et al. Detecting protein function and protein–protein interaction from genome sequences. Science 1999;285:751–753.
Pellegrini M, Marcotte EM, Thompson MJ, et al. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc Natl Acad Sci USA 1999;96:4285–4288.
Dandekar T, Snel B, Huynen M, et al. Conservation of gene order: a fingerprint of proteins that physically interact. Trends Biochem Sci 1998;23:324–328.
Elston CW, Ellis IO . Pathological prognostic factors in breast cancer I. The value of histological grade in breast cancer: experience from a large study with long-term follow up. Histopathology 1991;19:403–410.
Chasle J, Delozier T, Denoux Y, et al. Immunohistochemical study of cell cycle regulatory proteins in intraductal breast carcinomas—a preliminary study. Eur J Cancer 2003;39:1363–1369.
Buxant F, Anaf V, Simon P, et al. Ki-67 immunostaining activity is higher in positive axillary lymph nodes than the primary breast tumor. Breast Cancer Res Treat 2002;75:1–3.
Assersohn L, Salter J, Powles TJ, et al. Studies of the potential utility of Ki67 as a predictive molecular marker of clinical response in primary breast cancer. Breast Cancer Res Treat 2003;82:113–123.
Molino A, Pedersini R, Micciolo R, et al. Relationship between the thymidine labelling and Ki-67 proliferative indices in 126 breast cancer patients. Appl Immunohistochem Mol Morphol 2002;10:304–309.
Talley LI, Grizzle WE, Waterbor JW, et al. Hormone receptors and proliferation in breast carcinomas of equivalent histologic grades in pre- and postmenopausal women. Int J Cancer 2002;98:118–127.
Boland GP, Butt IS, Prasad R, et al. COX-2 expression is associated with an aggressive phenotype in ductal carcinoma in situ. Br J Cancer 2004;90:423–429.
Martinez-Arribas F, Nunez MJ, Piqueras V, et al. Flow cytometry vs Ki67 labelling index in breast cancer: a prospective evaluation of 181 cases. Anticancer Res 2002;22:295–298.
Weidner N, Moore II DH, Vartanian R . Correlation of Ki-67 antigen expression with mitotic figure index and tumor grade in breast carcinomas using the novel ‘paraffin’-reactive MIB1 antibody. Hum Pathol 1994; 25:337–342.
Leek RD, Kaklamanis L, Pezzella F, et al. bcl2 in normal human breast and carcinoma, association with oestrogen receptor-positive, epidermal growth factor receptor-negative tumors and in situ cancer. Br J Cancer 1994;69:135–139.
Borner C . Diminished cell proliferation associated with the death-protective activity of Bcl-2. J Biol Chem 1996;272:12695–12698.
Huang DCS, O'Reilly LA, Strasser A, et al. The antiapoptosis function of Bcl-2 can be genetically separated from its inhibitory effect on cell cycle entry. EMBO J 1997;16:4628–4638.
Hoetelmans RWM, Van de Velde CJH, Van Direndonck JH . The presence of 19-kDa Bcl-2 in dividing cells. Cell Prolif 2003;36:293–306.
O'Reilly L, Huang DCS, Strasseer A . The cell death inhibitor bcl-2 and its homologues influence control of cell cycle entry. EMBO J 1996;15:6979–6990.
Han EKH, Begemann M, Sgambato A, et al. Increased expression of cyclin D1 in a murine mammary epithelial cell line induces p27kip1, inhibits growth, and enhances apoptosis. Cell Growth Differ 1996;7: 699–710.
Eischen CM, Woo D, Roussel MF, et al. Apoptosis triggered by myc-induced suppression of Bcl-X(L) or Bcl-2 is bypassed during lymphomagenesis. Mol Cell Biol 2001;21:5063–5070.
Nilsson JA, Cleveland JI . Myc pathways provoking cell suicide and cancer. Oncogene 2003;22:9007–9021.
Visscher DW, Sarker F, Tabaczka P, Crissman J . Clinicopathologic analysis of bcl-2 immunostaining in breast carcinoma. Mod Pathol 1996;9:642–646.
Su GH, Song JJ, Repasky EA, et al. Mutation rate of MAP2K4/MKK4 in breast carcinoma. Hum Mutat 2002;19:81.
Griffith TS, Broghammer EL . Suppression of tumor growth following intralesional therapy with TRAIL recombinant adenovirus. Mol Ther 2001;4:257–266.
Shankar S, Srivastava RK . Enhancement of therapeutic potential of TRAIL by cancer chemotherapy and irradiation: mechanisms and clinical implications. Drug Resist Updat 2004;7:139–156.
Acknowledgements
This study was supported by grants from the Singapore Cancer Society and the Singapore Cancer Syndicate (SCS-MS0004).
Author information
Authors and Affiliations
Corresponding author
Additional information
Parts of this paper were presented at the 93rd Annual Meeting of the United States and Canadian Academy of Pathology, Vancouver, BC, Canada, March 6–12, 2004.
Rights and permissions
About this article
Cite this article
Tan, PH., Bay, BH., Yip, G. et al. Immunohistochemical detection of Ki67 in breast cancer correlates with transcriptional regulation of genes related to apoptosis and cell death. Mod Pathol 18, 374–381 (2005). https://doi.org/10.1038/modpathol.3800254
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.3800254
Keywords
This article is cited by
-
Subtype-specific network organization of molecular complexes in breast cancer
Journal of Biosciences (2023)
-
Accuracy of p53 and ki-67 in the graduation of phyllodes tumor, a model for practical application
Surgical and Experimental Pathology (2020)
-
Isolation and characterization of the primary epithelial breast cancer cells and the adjacent normal epithelial cells from Iranian women’s breast cancer tumors
Cytotechnology (2018)
-
Prognostic Value of Ki-67 in Breast Carcinoma: Tissue Microarray Method Versus Whole Section Analysis- Potentials and Pitfalls
Pathology & Oncology Research (2015)
-
[18F]FDG and [18F]FLT positron emission tomography imaging following treatment with belinostat in human ovary cancer xenografts in mice
BMC Cancer (2013)