Abstract
Synovial sarcomas usually occur in the soft tissues of the extremities of adolescents and middle-aged patients, in the vicinity of large joints. We present a patient with a synovial sarcoma of the left atrium and ventricle, which is an extremely rare location. Diagnosis was confirmed by reverse transcriptase polymerase chain reaction (RT-PCR), showing the t(X;18) fusion transcript. With a multicolor COmbined Binary RAtio labeling Fluorescence In Situ Hybridization (COBRA-FISH) technique, a complex karyotype evolved with identification of derivative chromosomes with multiplex rearrangements. This underscores the importance of molecular analyis of spindle cell tumors in unusual locations. Moreover, it shows that the presumed diagnostic translocation t(X;18) can be embedded in a sequence of other chromosomal rearrangements of which the function is as yet unknown.
Similar content being viewed by others
Main
Primary tumors of the heart and pericardium are rare, and about 65–90% of them are benign, of which most are myxomas. Of the primary sarcomas, angiosarcoma and malignant fibrous histiocytoma (MFH) are the most common, whereas synovial sarcoma (SS) accounts for only 3%.1 In the literature, about 13 cases of primary SS have been described (Table 1), most of which are restricted to histological and immunohistochemical data. In this report, we present a case of a cardiac SS with cytogenetic analysis using a multicolor COmbined Binary RAtio labeling Fluorescence In Situ Hybridization (COBRA-FISH) method and with reverse transcriptase-polymerase chain reaction (RT-PCR) analysis of the t(X;18) SYT-SSX fusion gene product.
Materials and methods
Case Report
A 42-year-old male complained of retrosternal pain, shortness of breath and fever. Radiological analysis by X-ray showed a mass adjacent to the left atrium extending to the annulus of the mitral valve and posterior wall of the left ventricle with pericardial fluid (Figure 1a). Thoracotomy was performed, but only a resection of the atrial part of the tumor was possible. Investigation by frozen section during surgery showed a mesenchymal malignancy. Further, computed tomography (CT) analysis (Figure 1b) showed multiple lung metastases and probable liver metastases. In consultation with the patient, no further therapy was instituted. The patient died of metastatic disease 1 month after the operation. Autopsy was not permitted.
(a) Posteroanterior chest radiograph (obtained before surgery) reveals a large mass (T) adjacent to the left ventricle. (b) Contrast-enhanced CT at the level of the left ventricle (performed after initial surgery) revealed a hypodense tumor mass (T) of max. 6 cm, at this height adherent to the dorsal myocardium of the left ventricle (V). Note pericardial effusion (*). In the same CT-procedure, multiple lung and liver metastases were found.
Immunohistochemistry
Immunohistochemistry was performed as described earlier in a two-step staining method by a Techmate 500 immunostainer (DAKO).2 Antibodies used were directed against keratin proteins AE1-AE3 (Boehringer), Vimentin (DAKO), SM-actin (Progen), MS-actin (DAKO), Desmin (DAKO), S100 (DAKO), neurofilaments (Sanbio, Uden, the Netherlands), CD31 (DAKO), CD34 (Neomarkers), epithelial membrane antigen (DAKO), Ber-Ep 4 (DAKO), Calretinin (Novocastra), CD56 (Becton Dickinson), BCL-2 (DAKO), and E-cadherin (Bionovo, Duiven, Netherlands).
Tumor Sample and Chromosome Preparations
A fresh sample of tumor tissue was mechanically minced and subsequently inoculated into a culture flask containing 5 ml of RPMI 1640 medium 1 mg/ml collagenase 1A (Sigma-Aldrich) and 1 mg/ml dispase (GIBCO) to enzymatically disaggregate cells. After 16 h of treatment, released tumor cells were washed and cultured in RPMI 1640 medium, supplemented with 10% fetal calf serum and penicillin/streptomycin. After 5 days of incubation, metaphase cells were harvested using colcemid (20 ng/ml) and incubated for 2 h. After trypsinization and hypotonic treatment (0.075 M KCl for 12 min) cells were fixed by three changed fixation steps using methanol/glacial acetic acid (3:1 v/v). The cell suspension was dropped and selected slides were used for FISH analysis.
Molecular Cytogenetic Analysis
Slides with metaphase chromosomes were hybridized using a multicolor FISH approach named COBRA-FISH. 48-color FISH staining every chromosome-arm in a different color combination, digital imaging and analysis were performed as previously described by our group.3, 4, 5 Hybridizations with individual libraries labeled with single fluorochromes were used to confirm the detected rearrangements. Breakpoints were assigned using inverted 4,6-diamidino-2-phenylindole (DAPI) counterstained images of the chromosomes. Karyotypes have been described according to the International System for Human Cytogenetic Nomenclature (ISCN, 1995).6
Owing to the involvement of chromosome 22q12, known to be a breakpoint in the t(11;22)(q24;q12) in Ewing's sarcoma, a cosmid probe set covering the EWS region on chromosome 22 was used. Two cosmid probes covering the 5′ (G9) and 3′ (F7) parts of the breakpoint regions7 were labeled with fluorescein isothiocyanate (FITC) or Cy3 d-UTPs using random prime labeling kit (Invitrogen) according to the manufacturer's instructions. On normal chromosome 22, the simultaneous hybridization of the two differentially labeled cosmids results in a colocalization of the green and red signals. In the case of a translocation involving the EWS region, the two probes do not cohybridize and the green and red signals locate to different chromosomes.
RT-PCR
RNA isolation, reverse transcription, PCR, and gel electrophoresis were performed as described earlier.8 In short, RNA was extracted from fresh frozen tumor tissue with TRIzol (Gibco BRL Life Technologies, Gaithersburg, MD, USA). RNA was reversely transcribed in a mix containing 2 μg RNA and 0.2 μl AMV-reverse transcriptase (Boehringer Mannheim, Germany). Amplification by PCR was carried out using 1 U Ampli-Taq polymerase (Perkin-Elmer Cetus, Norwalk, CT, USA) in a Perkin-Elmer Thermal cycler. A total of 30 cycles were performed of 30 s at 94°C (denaturation), 60 s at 68°C (annealing) and 60 s at 72°C (elongation). SSX- (5′-CACTTGCTATGCACCTGATG-3′, backward) and SYT- (5′-CAACAGCAAGATGCATACCA-3′, forward) primers were used. Hypoxanthine Phosphoribosyl Transferase (HPRT) complementary DNA (cDNA) was amplified with primers hum1 and hum2. As a positive control, cDNA of a cytogenetically confirmed t(X;18)-positive patient was used, and as a negative control H2O was used. PCR products of 10 μl were subjected to electrophoresis in a 1.2% agarose gel, and stained with ethidium bromide.
Results
Histology and Immunohistochemistry
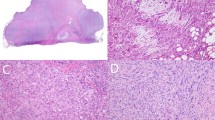
Multiple tissue blocks from the resected tumor material were available. These showed a tumor infiltrating pre-existing cardiac tissue, consisting of monomorphic spindle cells in a partly haphazard, partly storiform pattern with indistinct cell borders. The intervening areas showed focal myxoid changes and focal necrosis (Figure 2). Tumor nuclei had small, sometimes multiple chromocenters or nucleoli, and an average of 4 mitoses per 2 mm2 were seen. No epithelial differentiation was observed, a Gomori stain revealed no ‘nests’ of epithelium. Immunohistochemistry showed clusters and isolated pankeratin-positive tumor cells; CD56 and BCL-2 were strongly positive; epithelial membrane antigen and calretinin were heterogeneously, focally positive. SM-actin, MS-actin, desmin, S-100, neurofilaments, CD31, CD34, Ber-Ep4 and E-cadherin were negative.
(a) Overview of spindle cell tumor infiltrating myocardial tissue (lower left). No epithelial differentiation is seen (hematoxylin and eosin, × 50). (b) Myxoid changes of the tumor can be seen at the lower left (× 100). (c) Plump spindle nuclei with moderate anisokaryosis, coarse chromatin, and multiple mitotic figures (× 200). (d) Tumor shows intense and diffuse positivity for pankeratin staining (× 100).
Molecular Cytogenetic Analysis
Multicolor FISH-based karyotyping was conducted using a 48-color FISH allowing the identification of all chromosomes and short and long arms of individual chromosomes. A representative karyotype image is shown in Figure 3. The following near-triploid karyotype was ascertained: ∼61,Y,der(X)t(X;18)(p1;p1)x2,der(1)t(1;8)(q10;q10),der(4)t(4;14)(p14;q11∼12),+5,−6,del(7)(p13∼14),−9,−10,−11,der(11)t(11;12)(q10;p10)x2,der(12)t(X;12)(p;q),−13,−14,−14,−16,+17,−18,del(18)(p10),−20,−21,der(22)t(12;22)(q12;q12)t(11;12)(q24∼25;q24),der(22)t(5;22)(p10;p11)x2[cp25].
Representative karyogram of a tumor cell after COBRA-FISH hybridization. All rearranged chromosomes have been validated by using combinations of wcp probes resulting in the following near-triploid karyotype: ∼61,Y,der(X)t(X;18)(p1;p1)x2,der(1)t(1;8)(q10;q10),der(4)t(4;14)(p14;q11∼12),+5,−6, del(7)(p13∼14),−9,−10,−11, der(11)t(11;12)(q10;p10)x2,der(12)t(X;12)(p;q),−13,−14,−14,−16,+17,−18,del(18)(p10),−20,−21,der(22)t(12;22)(q12;q12)t(11;12)(q24∼25;q24), der(22)t(5;22)(p10;p11)x2[cp25]. Hybridizations with chromosome X and 18 wcp probes verified the lack of the der(18)t(X;18). Inset shows the result of EWS region-specific cosmid (red, green) and wcp22 (blue) hybridization superimposed on DAPI image (gray). Colocalization of red and green signals proved that the breakpoint on chromosome 22 is distal of EWS region.
Whole chromosome painting probe (Wcp probe) hybridizations were used with combinations of probe sets corresponding to the rearrangements. Hybridizations with chromosome X and 18 wcp probes verified the lack of the der(18)t(X;18). A cosmid probe set covering the EWS gene region on chromosome 22 showed no involvement of this gene.
RT-PCR
Detection of the SYT-SSX fusion transcript by RT-PCR showed a fusion product in the expected range of 585 base pairs, at the same level as the positive control (Figure 4). This was confirmed by hybridization with a SYT/SSX probe (not shown).
RT-PCR analysis. Electrophoresis gel shows 100 bp size marker (L), HPRT housekeeping gene control (H), extracted RNA of the patient's tumor (P), RNA of SS with cytogenetically proven t(X;18)(p11;p11) (+), and H2O as negative control (−). Patient's cDNA and positive control show a product in the expected range of 585 bp.
Discussion
SS of the heart are extremely rare. Patients generally present with complaints of dyspnea, fatigue, chest pain, or congestive heart failure, sometimes with syncope or ischemic attacks due to tumor emboli.9, 10, 11, 12 Most tumors are located in the right heart (see Table 1), and males outweigh females by about 2.5 to 1. As can be seen in Table 1, patients with cardiac SS have an extremely adverse prognosis, as eight of 11 patients with known follow-up died shortly after diagnosis and therapy, and the three ‘survivors’ have been followed for no more than 12 months. Of course, the main reason for this is the location of the tumor, making radical surgery impossible for most patients.
In the clinical work-up, cardiac myxoma may be a differential diagnosis, whereas histologically malignancy is usually easily established. The main differential diagnoses may include mesothelioma (especially in the biphasic variant), malignant peripheral nerve sheath tumor, or fibrosarcoma (in the case of a monophasic spindle cell variant).1, 13 As SS may have areas of myxoid differentiation, in selected biopsies a myxoma or myxoid sarcoma may be considered as well. Differentiation from mesothelioma may be difficult at the histological level, but distinction may be possible by use of immunohistochemical panels.
In some cases, however, the difference can only be made upon molecular analysis, especially in this case of a spindle cell tumor in an unusual location. The hallmark of diagnosis of SS is the detection of the t(X;18)(p11.2;q11.2), which is present in more than 90% of SS.14 This translocation leads to the fusion of the 5′ end of the SYT-gene at 18q11.2 to the 3′ end of either SSX1 or SSX2, closely related at Xp11.2.15 It was recently argued that molecular testing for SS is not necessary when diagnosis is certain or probable on the basis of clinical, histologic and immunohistochemical data, because a large majority of these tumors in the tested series showed the t(X;18).16 We propose that especially at uncommon locations like in our patient, molecular confirmation of SS by cytogenetics or RT-PCR remains a valuable adjunct in diagnosis.
There seems to be no correlation between karyotype and histologic subtype or prognosis,14, 17 and the impact of SYT-SSX fusion type remains a matter of debate. Ladanyi et al18 showed in a large series of patients that in the subgroup of localized disease, patients with a SYT-SSX2 translocation had significantly better survival than patients with SYT-SSX1 tumors. No cardiac tumors were included in this series of 243 patients. In another large series (182 patients), SYT-SSX fusion type was claimed not to be a prognostic factor.19 In both studies, tumor histology (monophasic vs biphasic) had no impact on survival. How the SYT-SSX chimeric proteins are exactly related to tumorigenesis in SS remains to be established, but the fusion protein is thought to be involved in the transcriptional deregulation of specific target genes.20
In about one-third of SS, the t(X;18)(p11.2;q11.2) is the sole cytogenetic abnormality, and the majority has a near-diploid karyotype. Complex and additional abnormalities are relatively more frequent in local recurrences and metastases.14 Complex t(X;18) translocations involving three or more chromosomes constitute less than 10% of reported SS cases.14 In the Mitelman database of chromosome aberrations in cancer,21 approximately one-quarter of 180 SS does not show a near-diploid karyotype, and around one-half shows additional translocations. However, these data may be seriously biased as many ‘simple’ karyotypic cases—unless at uncommon locations—may not have been published. The degree of aneuploidy and number of additional translocations that was found in our case of primary SS is rather uncommon; only around 15 cases in the Mitelman database show this degree of aneuploidy, and only 20 cases do have more than two additional translocations. The complex karyotype with multiple additional translocations that was found in our case of primary SS is rather uncommon. None of the additional translocations were found to be specific for any kind of tumor. Because of the involvement of chromosome 22q12, a cosmid probe set covering the EWS region was used. There was no involvement of this region. Of the three other cardiac SS cases for which cytogenetic data have been presented, one had a complex t(X;18) involving chromosome 7;22 in the remaining two, only the existence of the t(X;18) was mentioned.23, 24 In two papers, the SYT-SSX fusion gene has been presented by RT-PCR.25, 26 This leaves restricted data for comparison of the cardiac cases in particular. The detection and description of the complex karyotype was facilitated by the use of multicolor FISH. The use of (multicolor-)FISH-based identification of chromosomes is particularly useful in cases with complex karyotypes or with changes that would escape detection based solely on banding pattern changes of the rearranged chromosome segments.27 In our case, the use of the multicolor FISH approach—that allows the separate identification of short and long arms of all chromosomes—resulted in the identification of derivative chromosomes with multiplex rearrangements in a single experiment.
In conclusion, we present a patient with SS of the left atrium and ventricle with confirmation of the t(X;18) by RT-PCR and multicolor COBRA-FISH, the latter establishing a complex karyotype.
References
Burke AP, Virmani R . Primary cardiac sarcomas. In: Burke AP, Virmani R (eds). Tumors of the Heart and Great Vessels. Atlas of Tumor Pathology, AFIP, fasc. 16. 3rd ed. Armed Forces Institute of Pathology: Washington, 1996, pp 127–170.
Hazelbag HM, Laforga JB, Roels HJL, et al. Dedifferentiated adamantinoma with revertant mesenchymal phenotype. Am J Surg Pathol 2003;27:1530–1537.
Tanke HJ, Wiegant J, Van Gijlswijk RPM, et al. New strategy for multi-colour fluorescence in situ hybrydisation: COBRA: COmbined Binary RAtio labelling. Eur J Hum Genet 1999;7:2–11.
Wiegant J, Bezrookove V, Rosenberg C, et al. Differentially painting human chromosome arms with combined binary ratio-labeling fluorescence in situ hybridization. Genome Res 2000;10:861–865.
Szuhai K, Bezrookove V, Wiegant J, et al. Simultaneous molecular karyotyping and mapping of viral DNA integration sites by 25-color COBRA-FISH. Genes Chromosomes Cancer 2000;28:92–97.
ISCN. An International System for Human Cytogenetic Nomenclature. S Karger: Basel, 1995.
Zucman J, Delattre O, Desmaze C, et al. Cloning and characterization of the Ewing's sarcoma and peripheral neuroepithelioma t(11;22) translocation breakpoints. Genes Chromosomes Cancer 1992;5:271–277.
Bovee JVMG, Devilee P, Cornelisse CJ, et al. Identification of an EWS-pseudogene using translocation detection by RT-PCR in Ewing's sarcoma. Biochem Biophys Res Commun 1995;213:1051–1060.
Donsbeck AV, Ranchere D, Coindre JM, et al. Primary cardiac sarcomas: an immunohistochemical and grading study with long-term follow-up of 24 cases. Histopathology 1999;34:295–304.
Casselman FP, Gillinov AM, Kasirajan V, et al. Primary synovial sarcoma of the left heart. Ann Thorac Surg 1999;68:2329–2331.
Bittira B, Tsang J, Huynh T, et al. Primary right atrial synovial sarcoma manifesting as transient ischemic attacks. Ann Thorac Surg 2000;69:1949–1951.
Nicholson AG, Rigby M, Lincoln C, et al. Synovial sarcoma of the heart. Histopathology 1997;30:349–352.
Pauwels P, Dal Cin P, Sciot R, et al. Primary malignant peripheral nerve sheath tumour of the heart. Histopathology 1999;34:56–59.
Sandberg AA, Bridge JA . Updates on the cytogenetics and molecular genetics of bone and soft tissue tumors. Synovial sarcoma. Cancer Genet Cytogenet 2002;133:1–23.
Clark J, Rocques PJ, Crew AJ, et al. Identification of novel genes, SYT and SSX involved in the t(X;18)(p11.2;q11.2) translocation found in human synovial sarcoma. Nat Genet 1994;7:502–508.
Coindre JM, Pelmus M, Hostein I, et al. Should molecular testing be required for diagnosing synovial sarcoma? A prospective study of 204 cases. Cancer 2003;98:2700–2707.
Fletcher CDM, Dal Cin P, De Wever I, et al. Correlation between clinicopathological features and karyotype in spindle cell sarcomas. Am J Pathol 1999;154:1841–1847.
Ladanyi M, Antonescu CR, Leung DH, et al. Impact of SYT-SSX fusion type on the clinical behavior of synovial sarcoma: a multi-institutional retrospective study of 243 patients. Cancer Res 2002;62:135–140.
Guillou L, Benhattar J, Terrier P, et al. SYT-SSX fusion type is not a prognostic factor in synovial sarcoma (SS) patients. A multi-institutional study of 182 cases. Mod Pathol 2003;16:13A.
Ladanyi M . Fusions of the SYT and SSX genes in synovial sarcoma. Oncogene 2001;20:5755–5762.
Mitelman F, Johansson B, Mertens F . Mitelman database of chromosome aberrations in cancer. http://cgap.nci.nih.gov/chromosomes 2003.
Iyengar V, Lineberger AS, Kerman S, et al. Synovial sarcoma of the heart—correlation with cytogenetic findings. Arch Pathol Lab Med 1995;119:1080–1082.
Karn CM, Socinski MA, Fletcher JA, et al. Cardiac synovial sarcoma with translocation (X 18) associated with asbestos exposure. Cancer 1994;73:74–78.
McGilbray TT, Schulz TK . Primary cardiac synovial sarcoma. Lancet Oncol 2003;4:283.
Oizumi S, Igarashi K, Takenaka T, et al. Primary pericardial synovial sarcoma with detection of the chimeric transcript SYT-SSX. Jpn Circul J—Engl Ed 1999;63:330–332.
Al Rajhi N, Husain S, Coupland R, et al. Primary pericardial synovial sarcoma: a case report and literature review. J Surg Oncol 1999;70:194–198.
Mathew S, Dalton J, Riedley S, et al. Complex t(X;18)(p11.2;q11.2) with a pericentric inversion of the X chromosome in an adolescent boy with synovial sarcoma. Cancer Genet Cytogenet 2002;132:136–140.
McAllister HA, Fenoglio JJ . Malignant tumors of the heart and pericardium. In: Tumors of the Cardiovascular System (fasc. 15). AFIP Atlas of Tumor Pathology, Second Series. AFIP: Bethesda, 1978, p 108.
Sheffield EA, Corrin B, Addis BJ, et al. Synovial sarcoma of the heart arising from a so-called mesothelioma of the atrioventricular node. Histopathology 1988;12:191–202.
Siebenmann R, Jenni R, Makek M, et al. Primary synovial sarcoma of the heart treated by heart transplantation. J Thorac Cardiovasc Surg 1990;99:567–568.
Burke AP, Cowan D, Virmani R . Primary sarcomas of the heart. Cancer 1992;69:387–395.
Acknowledgements
This project was financially supported by a grant from the Dutch Cancer Foundation (KWF2001-2526). We thank A de Roos, MD, PhD, for critical review of the radiology and E Kerkhof, MSc, for performing the RT-PCR.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hazelbag, H., Szuhai, K., Tanke, H. et al. Primary synovial sarcoma of the heart: a cytogenetic and molecular genetic analysis combining RT-PCR and COBRA-FISH of a case with a complex karyotype. Mod Pathol 17, 1434–1439 (2004). https://doi.org/10.1038/modpathol.3800200
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.3800200
Keywords
This article is cited by
-
Laparoscopic intragastric resection of gastric synovial sarcoma: report of the first ever case with video demonstration
World Journal of Surgical Oncology (2021)
-
Primary Synovial Sarcoma (SS) of the digestive system: a molecular and clinicopathological study of fifteen cases
Clinical Sarcoma Research (2015)
-
A balanced t(5;17) (p15;q22-23) in chondroblastoma: frequency of the re-arrangement and analysis of the candidate genes
BMC Cancer (2009)
-
COBRA: combined binary ratio labeling of nucleic-acid probes for multi-color fluorescence in situ hybridization karyotyping
Nature Protocols (2006)
-
Synovial sarcoma with radiological appearances of primitive neuroectodermal tumour/Ewing sarcoma: differentiation by molecular genetic studies
Skeletal Radiology (2006)