Abstract
Macrolide-resistant Helicobacter (H.) pylori represent an increasing therapeutic problem. Macrolide resistance is usually determined phenotypically in vitro with methods such as E-test or agar dilution test. A prerequisite for those tests, however, is bacterial culture that is not routinely set up in the course of gastroscopy. In contrast, formalin-fixed, paraffin-embedded biopsies are regularly available from patients who have undergone gastroscopy. In such biopsies macrolide-resistant H. pylori can be detected by the genotype-based technique of fluorescence in situ hybridization (FISH). Experience gained by this new method, however, is still extremely limited, especially in formalin-fixed tissue. Therefore, we retrospectively investigated formalin-fixed, paraffin-embedded biopsy specimens by FISH in 104 patients suffering from therapy-resistant H. pylori gastritis. To test the accuracy of FISH, we initially examined specimens from 53 patients for whom results of the E-test were available. Next we analyzed biopsies from another 51 patients that had been selected since phenotypical resistance testing had failed despite documented culturing attempts. In all 104 patients, H. pylori was detected by FISH and could thus be investigated for macrolide resistance. Overall, macrolide-resistant bacteria were found in 71 patients (68.3%). In 49 of 53 patients (92.4%), FISH and E-test returned identical results (no significant discordance according to McNemar's χ2-test). Taken together, our study demonstrates that FISH is a highly sensitive and reliable method for detecting macrolide-resistant H. pylori in formalin-fixed, paraffin-embedded biopsy specimens, which represents the routine method of processing tissue obtained upon gastroscopy.
Similar content being viewed by others
Main
Helicobacter (H.) pylori is a bacterial pathogen associated with a range of gastric disorders, such as chronic gastritis, peptic ulcer, mucosa-associated lymphoid tissue (MALT) lymphoma and adenocarcinoma of the stomach.1, 2, 3, 4 A triple-therapy regimen comprising a proton pump inhibitor in combination with the macrolide clarithromycin and amoxicillin or metronidazole is recommended as first-line treatment of H. pylori infection in Europe.5, 6 Bacterial resistance to antimicrobial agents is currently considered as an increasing problem in the treatment of infectious diseases in general.7 Resistance of H. pylori to clarithromycin is regarded as a particular dilemma, since this drug is a part of both established therapy regimen. Thus, macrolide resistance is a frequent cause for failure of H. pylori eradication therapy8 and secondary clarithromycin resistance rates in patients with a history of failed eradication attempts frequently exceed 50%,9, 10, 11 but also in the general population the prevalence of macrolide-resistant H. pylori has been increasing over the years, primarily in the industrial countries.12 Primary clarithromycin resistance rates reach up to 10% in Europe.13
Macrolide resistance of H. pylori is typically tested phenotypically in vitro, and the results are not available before 48–96 h after inoculation of the agar plates. Furthermore, culturing of H. pylori necessary for the phenotypic resistance testing method may be problematic, in particular after extended transportation times.14 Therefore, molecular methods for detection of genomic mutations associated with antibiotic resistance are attracting expanding attention.15 In clinical H. pylori isolates, macrolide resistance is determined by mutations of the 23S rRNA.16, 17 Trebesius et al18 developed a test that applies FISH to identify the most common of these mutations. This test can also be employed on formalin-fixed paraffin-embedded tissue samples.
The experience gained with this new method under routine clinical settings, however, is still very limited. To our knowledge, from formalin-fixed samples such experience has only been reported from 17H. pylori-infected patients, of whom only five were infected with a bacterial strain resistant to macrolides.18 This prompted us to perform a retrospective study on formalin-fixed, paraffin-embedded gastric biopsies obtained from a total of 104 patients with therapy-resistant H. pylori infection and to compare the results generated by FISH with those gained by culture-based testing.
Material and methods
Patient Selection, Histological and Microbiological Evaluation
In centers throughout Germany, (gastroenterologists in private practice, communal hospitals and university departments), H. pylori-infected patients suffering from therapy-resistant H. pylori infection were screened for recruitment to a prospective therapy study.19 All patients were diagnosed with persisting H. pylori gastritis despite two or more documented antibiotic eradication attempts. Moreover, in at least one of those eradication tries, patients had to have been treated with a therapy regimen containing clarithromycin.
For histological investigation, two biopsies were taken from the antrum and another two from the corpus, immediately fixed in formalin, embedded in paraffin, sectioned (4 μm slice thickness) and dehydrated in an increasing series of ethanol/xylol. One section each was stained with hematoxylin and eosin (H&E) or with Warthin Starry silver stain (WS). The diagnosis of gastritis was established in accordance with the updated Sydney system.20
For microbiological culture, two further biopsies each were obtained from the antrum and corpus, immediately placed in Portagerm pylori medium (Biomérieux, Nürtingen, Germany), and transported in a precooled transport container (Sarstedt, Darmstadt, Germany) to a university department (Regensburg or Dresden) for microbiological investigation.14 At both the departments of clinical microbiology, primary H. pylori culture and antimicrobial susceptibility testing was performed in an identical manner using anaerobic jars that contained an atmosphere of 9% CO2, 11% O2 and 80% N2 incubated at 36°C. Susceptibility testing was performed using the E-test and a minimal inhibitory concentration ≥0.5 μg/ml was regarded as clarithromycin resistant.19
Identification of H. pylori and Determination of Macrolide Resistance using FISH
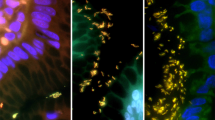
The same formalin-fixed paraffin-embedded gastric biopsies that had been used for histopathological assessment of H. pylori gastritis were evaluated by FISH. Biopsies were sectioned and dehydrated as described above. Then sections were air-dried and hybridized using the commercially available test system CreaFast H. pylori Combi Kit (Creatogen, Augsburg, Germany) according to the manufacturer's instructions. This test kit that is now distributed by Seapro Diagnostics Ltd, Liverpool, UK, labels H. pylori with the green fluorescent stain, fluorescein, and resistant H. pylori additionally with Cy3-red. Following hybridization, the sections were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI) (1 μg/ml; Sigma, Heidelberg, Germany). Slides were inspected with a BX-60 microscope (Olympus, Hamburg, Germany) equipped with a standard fluorescence filter set. Vegetative H. pylori forms were localized on the basis of the blue fluorescent DAPI staining and identified using the filter for green fluoescence. Macrolide resistant H. pylori were detected with the aid of the Cy3-specific filter (see Figure 1).
Detection of macrolide-resistant H. pylori by FISH in gastric biopsy specimens. (a) Identification of vegetative forms (marked with “ → ”) due to hybridization with H. pylori-specific, fluorescein-labelled probe (green fluorescence). (b) Detection of bacterial resistance against macrolides due to hybridization with a Cy3-labelled mixture of probes specific for the most frequent 23S rRNA mutations associated with macrolide resistance (same section as (a); red fluorescence). (c) Simultaneous colonization with macrolide-sensitive (marked with “ → ”) and resistant (marked with “ ⇒ ”) H. pylori. Note the yellow appearance of resistant bacteria that results from an additive mixture of red and green fluorescence.
Statistical Analysis
The discordance between the results of phenotypic and genotypic resistance testing was compared using McNemar's χ2-test with an automatic on-line calculation (http://graphpad.com/quickcalcs/McNemar1.cfm). 95% confidence intervals were also calculated on-line (http://graphpad.com/quickcalcs/ConfInterval1.cfm).
Results
Initially, gastric biopsies obtained from 53 patients with histopathologically diagnosed H. pylori gastritis and successful phenotypic resistance testing were investigated by FISH. The bacteria were detected in all 53 patients, with 36 patients (67.9%) revealing the presence of clarithromycin-resistant bacteria. Concordance with the phenotypic resistance testing was thus 92.4%. In four patients, FISH and phenotypic resistance testing led to discrepant results: in the specimens obtained from two patients, macrolide-resistant H. pylori were detected by FISH, but not by the E-test, while the reverse was the case in two further patients (see Table 1). According to McNemar's χ2-test, the number of discrepant results did not reach statistical significance. Subsequent sections obtained from the four patients with differing results gained by the two detection methods were re-investigated by FISH: in one patient originally diagnosed as harboring only clarithromycin-sensitive H. pylori in the stomach, careful re-evaluation revealed a very small percentage (less than 1%) of macrolide-resistant bacteria, while in the other three cases the original findings were confirmed. From one of these patients who, according to FISH but not to E-test, was infected with macrolide-resistant H. pylori, additional biopsy material was available that had been obtained approximately 2 weeks prior to the material investigated originally. Investigation of this material by FISH identified macrolide-sensitive H. pylori only. Overall FISH proved to be an accurate method for the determination of H. pylori's resistance to macrolides in formalin-fixed, paraffin-embedded gastric biopsies.
In a next step we tested if FISH was able to detect clarithromycin resistance in patients in which cultural resistance testing had been intended but failed. For this, we selected formalin-fixed biopsies obtained from 51 patients in whom H. pylori gastritis had been diagnosed histologically but phenotypic resistance testing had been impossible due to culture failure or contamination of the cultures. In the biopsies of all these patients, FISH was able to detect H. pylori. Macrolide-resistant bacteria were found in 35 of these 51 patients (68.6%).
Taken together, H. pylori was successfully detected in specimens from all 104 patients investigated (detection rate 100%, 95% confidence interval: 96–100%), with a total of 71 patients (68.3%) showing colonization with clarithromycin-resistant organisms.
Discussion
Analyzing more than 100 H. pylori-infected patients, this study shows that FISH is a very reliable method for detecting macrolide resistance in formalin-fixed biopsy specimens. To our knowledge, only four previous investigations employed FISH for the detection of macrolide resistance.18, 21, 22, 23 Analyzing a large number of shock-frozen tissue sections Russman et al23 were able to show that FISH is a very sensitive method for the detection of H. pylori. Moreover, the same group and Feydt-Schmidt and co-workers provided convincing data that FISH is a very reliable technique for the detection of clarithromycin resistant H. pylori in shock-frozen tissue, bacterial cultures and biopsies that were processed for FISH within hours.21, 22, 23 Shock freezing of biopsies, setting up a H. pylori culture and immediate processing of samples for hybridization, however, are not routine procedures in everyday work of gastroscopical units. In contrast, formalin fixation of gastric biopsies represents a standard method. Therefore, we tested the reliability of FISH for the examination of macrolide resistance in formalin-fixed gastric biopsy specimens in a large collection of H. pylori-positive patients. Moreover, to include a relatively high percentage of macrolide-resistant H. pylori strains, only material from patients suffering from refractory H. pylori gastritis was analyzed. In this set-up, FISH proved a trustworthy and robust method for detecting macrolide resistance.
Since phenotypical resistance testing that represents the standard method for the determination of H. pylori's macrolide resistance continues to pose a problem in particular after lengthy transport times of the specimens,14 several genotype-based methods for the detection of macrolide resistance have been developed.18, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34 Most of these methods employ a mutation-specific PCR.24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34 Such methods, however, require the preparation of DNA and special molecular-biological equipment (eg real time PCR cycler). Furthermore, no reports on the effectiveness of these methods on formalin-fixed tissue specimens have been published yet. Therefore detection of macrolide resistance by FISH, which does not require special preparation of biopsy specimens and can be applied to formalin-fixed material, represents an approach of high interest. The results gained by this method are available within hours and the manpower requirements as well as the cost of the test kit (approximately 30 € per hybridization) are comparable to those of immunohistochemical investigations.
In a total of four of 53 patients, the results of testing with FISH were not in accordance with those of the E-test. In one of these patients, lack of experience of the investigator was the reason for the discrepancy, since investigating subsequently prepared slides from this patient who, according to the original FISH result, seemed to be infected with macrolide-sensitive H. pylori only, a very small percentage of clarithromycin-resistant bacteria (<1%) was found. Thus, the number of actually discrepant results was only three. It is a well-known fact that many patients harbor various H. pylori strains simultaneously.35 Moreover, each of these different strains can be resistant to a diverse spectrum of antibiotics.36 Therefore, a ‘sampling error’ of biopsies gained from patients with a mixed infection of resistant and susceptible strains is in our opinion the most probable explanation for the discrepant results between FISH and E-test. This view was further substantiated by a recent study that found 87.8% concordance established for two different laboratories separately investigating biopsies from identical patients.21 The influence of sampling error on the results of macrolide resistance testing by FISH is also suggested by our finding that in one of the three patients with discordant results of FISH and E-test analysis of additional formalin-fixed material by FISH returned the same results as did phenotypic resistance testing. Because of the impact of ‘false-sensitive’ results of resistance testing on H. pylori eradication, subsequent studies are required to establish biopsy regimens that minimize the influence of sampling error.
The overall macrolide resistance rate in the investigated group of patients with treatment-refractory H. pylori infection was 68.3%. This rate is in accordance with resistance rates of 54–71% reported in patients who had previously undergone unsuccessful triple-therapy eradication treatment.9, 10, 11 Owing to the high incidence of secondary macrolide resistance, in the treatment of therapy-refractory H. pylori infection, a therapeutic regimen containing clarithromycin has been recommended only when macrolide resistance can be excluded.5 For exclusion or detection of macrolide resistance, phenotypical resistance testing is currently regarded as the gold standard since this method is well-established and allows the simultaneous determination of H. pylori's resistance against various antibiotics including metronidazole, amoxicillin and tetracycline. There are, however, clinical situations in which genotype-based methods for the detection of H. pylori macrolide resistance such as FISH could help to save additional gastroscopic examinations. These situations certainly include cases when phenotypical resistance testing has been intended but failed due to negative culture or bacterial or fungal contamination of cultures. Also when bacterial culture has not been set up and histological examination reveals H. pylori infection in persons with an elevated risk of harboring clarithromycin-resistant bacteria, he or she can definitely benefit from the determination of macrolide resistance using genotype-based methods. Those patients comprise adults with a history of macrolide intake and H. pylori-infected children from areas or populations with a high resistance rate against macrolides.5 For those indications, determination of macrolide resistance by FISH is a rapid, simple and reliable method, as has been shown in the present study.
References
Blaser MJ . Hypotheses on the pathogenesis and natural history of Helicobacter pylori-induced inflammation. Gastroenterology 1992;102:720–727.
Blaser MJ, Perez-Perez GI, Kleanthous H, et al. Infection with Helicobacter pylori strains possessing cagA is associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res 1995;55:2111–2115.
Malfertheiner P, Bayerdorffer E, Diete U, et al. The GU-MACH study: the effect of 1-week omeprazole triple therapy on Helicobacter pylori infection in patients with gastric ulcer. Aliment Pharmacol Ther 1999;13:703–712.
Morgner A, Thiede C, Bayerdorffer E, et al. Long-term Follow-up of gastric MALT lymphoma after H. pylori Eradication. Curr Gastroenterol Rep 2001;3:516–522.
Malfertheiner P, Megraud F, O’Morain C, et al. Current concepts in the management of Helicobacter pylori infection—the Maastricht 2-2000 Consensus Report. Aliment Pharmacol Ther 2002;16:167–180.
Miehlke S, Bayerdorffer E, Graham DY . Treatment of Helicobacter pylori infection. Semin Gastrointest Dis 2001;12:167–179.
Cohen ML . Changing patterns of infectious disease. Nature 2000;406:762–767.
Megraud F . Resistance of Helicobacter pylori to antibiotics and its impact on treatment options. Drug Resist Updat 2001;4:178–186.
Ellenrieder V, Boeck W, Richter C, et al. Prevalence of resistance to clarithromycin and its clinical impact on the efficacy of Helicobacter pylori eradication. Scand J Gastroenterol 1999;34:750–756.
Heep M, Kist M, Strobel S, et al. Secondary resistance among 554 isolates of Helicobacter pylori after failure of therapy. Eur J Clin Microbiol Infect Dis 2000;19:538–541.
Peitz U, Sulliga M, Wolle K, et al. High rate of post-therapeutic resistance after failure of macrolide- nitroimidazole triple therapy to cure Helicobacter pylori infection: impact of two second-line therapies in a randomized study. Aliment Pharmacol Ther 2002;16:315–324.
Lahaie RG, Gaudreau C . Helicobacter pylori antibiotic resistance: trends over time. Can J Gastroenterol 2000;14:895–899.
Glupczynski Y, Megraud F, Lopez-Brea M, et al. European multicentre survey of in vitro antimicrobial resistance in Helicobacter pylori. Eur J Clin Microbiol Infect Dis 2001;20:820–823.
Heep M, Scheibl K, Degrell A, et al. Transport and storage of fresh and frozen gastric biopsy specimens for optimal recovery of Helicobacter pylori. J Clin Microbiol 1999;37:3764–3766.
Owen RJ . Molecular testing for antibiotic resistance in Helicobacter pylori. Gut 2002;50:285–289.
Debets-Ossenkopp YJ, Sparrius M, Kusters JG, et al. Mechanism of clarithromycin resistance in clinical isolates of Helicobacter pylori. FEMS Microbiol Lett 1996;142:37–42.
Versalovic J, Shortridge D, Kibler K, et al. Mutations in 23S rRNA are associated with clarithromycin resistance in Helicobacter pylori. Antimicrob Agents Chemother 1996;40:477–480.
Trebesius K, Panthel K, Strobel S, et al. Rapid and specific detection of Helicobacter pylori macrolide resistance in gastric tissue by fluorescent in situ hybridisation. Gut 2000;46:608–614.
Miehlke S, Kirsch C, Schneider-Brachert W, et al. A prospective, randomized study of quadruple therapy and high-dose dual therapy for treatment of Helicobacter pylori resistant to both metronidazole and clarithromycin. Helicobacter 2003;8:310–319.
Dixon MF, Genta RM, Yardley JH, et al. Classification and grading of gastritis. The Updated Sydney System. International Workshop on the Histopathology of Gastritis, Houston 1994. Am J Surg Pathol 1996;20:1161–1181.
Feydt-Schmidt A, Russmann H, Lehn N, et al. Fluorescence in situ hybridization vs epsilometer test for detection of clarithromycin-susceptible and clarithromycin-resistant Helicobacter pylori strains in gastric biopsies from children. Aliment Pharmacol Ther 2002;16:2073–2079.
Russmann H, Adler K, Haas R, et al. Rapid and accurate determination of genotypic clarithromycin resistance in cultured Helicobacter pylori by fluorescent in situ hybridization. J Clin Microbiol 2001;39:4142–4144.
Russmann H, Kempf VA, Koletzko S, et al. Comparison of fluorescent in situ hybridization and conventional culturing for detection of Helicobacter pylori in gastric biopsy specimens. J Clin Microbiol 2001;39:304–308.
Alarcon T, Domingo D, Prieto N, et al. PCR using 3′-mismatched primers to detect A2142C mutation in 23S rRNA conferring resistance to clarithromycin in Helicobacter pylori clinical isolates. J Clin Microbiol 2000;38:923–925.
Chisholm SA, Owen RJ, Teare EL, et al. PCR-based diagnosis of Helicobacter pylori infection and real-time determination of clarithromycin resistance directly from human gastric biopsy samples. J Clin Microbiol 2001;39:1217–1220.
Gibson JR, Saunders NA, Burke B, et al. Novel method for rapid determination of clarithromycin sensitivity in Helicobacter pylori. J Clin Microbiol 1999;37:3746–3748.
Maeda S, Yoshida H, Matsunaga, et al. Detection of clarithromycin-resistant Helicobacter pylori strains by a preferential homoduplex formation assay. J Clin Microbiol 2000;38:210–214.
Marais A, Monteiro L, Occhialini A, et al. Direct detection of Helicobacter pylori resistance to macrolides by a polymerase chain reaction/DNA enzyme immunoassay in gastric biopsy specimens. Gut 1999;44:463–467.
Matsuoka M, Yoshida Y, Hayakawa K, et al. Simultaneous colonisation of Helicobacter pylori with and without mutations in the 23S rRNA gene in patients with no history of clarithromycin exposure. Gut 1999;45:503–507.
Pina M, Occhialini A, Monteiro L, et al. Detection of point mutations associated with resistance of Helicobacter pylori to clarithromycin by hybridization in liquid phase. J Clin Microbiol 1998;36:3285–3290.
Ryan KA, van Doorn LJ, Moran AP, et al. Evaluation of clarithromycin resistance and cagA and vacA genotyping of Helicobacter pylori strains from the west of Ireland using line probe assays. J Clin Microbiol 2001;39:1978–1980.
Sevin E, Lamarque D, Delchier JC, et al. Co-detection of Helicobacter pylori and of its resistance to clarithromycin by PCR. FEMS Microbiol Lett 1998;165:369–372.
Stone GG, Shortridge D, Versalovic J, et al. A PCR-oligonucleotide ligation assay to determine the prevalence of 23S rRNA gene mutations in clarithromycin-resistant Helicobacter pylori. Antimicrob Agents Chemother 1997;41:712–714.
van Doorn LJ, Debets-Ossenkopp YJ, Marais A, et al. Rapid detection, by PCR and reverse hybridization, of mutations in the Helicobacter pylori 23S rRNA gene, associated with macrolide resistance. Antimicrob Agents Chemother 1999;43:1779–1782.
Taylor NS, Fox JG, Akopyants NS, et al. Long-term colonization with single and multiple strains of Helicobacter pylori assessed by DNA fingerprinting. J Clin Microbiol 1995;33:918–923.
Wang G, Jiang Q, Taylor DE . Genotypic characterization of clarithromycin-resistant and -susceptible Helicobacter pylori strains from the same patient demonstrates existence of two unrelated isolates. J Clin Microbiol 1998;36:2730–2731.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jüttner, S., Vieth, M., Miehlke, S. et al. Reliable detection of macrolide-resistant Helicobacter pylori via fluorescence in situ hybridization in formalin-fixed tissue. Mod Pathol 17, 684–689 (2004). https://doi.org/10.1038/modpathol.3800098
Received:
Revised:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.3800098
Keywords
This article is cited by
-
Sequencing-based methods and resources to study antimicrobial resistance
Nature Reviews Genetics (2019)
-
Helicobacter pylori Clarithromycin Resistance and Treatment Failure Are Common in the USA
Digestive Diseases and Sciences (2016)