Abstract
To identify the genes involved in cervical carcinogenesis, we applied the mRNA differential display (DD) method to analyze normal cervical tissue, cervical cancer, metastatic lymph node, and cervical cancer cell line. We cloned a 491-bp cDNA fragment, CC231, which was present in metastatic tissue and cervical cancer cell line, but absent in normal cervical and cervical cancer tissues. The 491 bp cDNA fragment has 98% homology to the previously published sequence, AAC-11 (antiapoptosis clone 11). The levels of AAC-11 mRNA expressions in nine normal cervical and nine primary cervical cancer tissues were low. Its expression was higher in three metastatic tissues and five cervical cancer cell lines (HeLa, CaSki, SiHa, CUMC-3, and CUMC-6). Invasion of matrigel and adhesion to laminin by AAC-11 transfected CUMC-6 cells were increased by approximately 2-fold and 4-fold, respectively. Northern blot analysis showed that matrix metalloproteinase (MMP)-2 and membrane type 1 MMP (MT1-MMP) genes were found to be expressed in high levels in AAC-11-transfected cancer cells. But MMP-2 and MT1-MMP were not expressed in cells transfected with vector alone or wild-type cells. AAC-11-transfected cells expressed an elevated level of MMP-2 protein as assessed by immunoblotting. On the contrary, tissue inhibitor of MMP (TIMP-2) expression was detectable in cells transfected with vector alone or wild-type cells, respectively. Its expression was undetectable in AAC-11 transfected cells. In cervical cancer cells transfected with AAC-11, the expression of β-catenin was up-regulated. These suggest that overexpressions of MMP-2 and MT1-MMP, loss of TIMP-2 expression, and up-regulation of β-catenin by AAC-11 transfection may contribute to the development of cervical cancer invasion. AAC-11 gene transfection increased cervical cancer cell colonization. The effect of AAC-11 on cultured cervical cancer cells was associated with antiapoptotic process. Approximately 50% of the AAC-11 transfected cells in serum-free medium died after 2 weeks, compared to 1 week for vector alone or wild-type cells. These results suggest that AAC-11 may serve as a candidate metastasis-related and apoptosis-inhibiting gene in human cervical cancer.
Similar content being viewed by others
Introduction
Cervical cancer is a classic model for the multistep nature of carcinogenesis, because it develops from well-defined precursors over a protracted period of time (Wright et al, 1994). Although molecular and epidemiological data clearly indicate that human papillomavirus (HPV) is the causal etiological factor for invasive cervical cancer (zur Hausen, 1991), HPV-associated carcinogenesis is an insufficient process, because only a small fraction of women who are infected with high-oncogenic-risk HPV types will ever develop invasive cervical cancer (Park et al, 1995). Altogether, especially in cervical cancers, the frequencies of oncogene activation (Choo et al, 1989; Kristensen et al, 1996) or tumor suppressor gene inactivation (Fujita et al, 1992) are lower than in other types of cancers. From this point of view, identification of another candidate oncogene or tumor suppressor gene that affects the cervical carcinogenesis is needed.
In this study, we applied the mRNA differential display (DD) method (Liang and Pardee, 1992) to find a new cancer-related gene in human cervical cancer. Finally, a candidate gene was identified and partially sequenced. We cloned a 491 bp cDNA fragment, CC231, which was present in metastatic lymph node tissue and cervical cancer cell line but absent in normal cervical and primary cervical cancer tissues. The 491 bp CC231 cDNA fragment has 98% homology to the previously published sequence, AAC-11 gene (accession number U83857), whose expression prevents apoptosis that occurs after growth factor deprivation (Tewari et al, 1997). BALB/c3T3 fibroblasts that were stably transfected with AAC-11 cDNA were viable in serum-free medium for up to 12 weeks.
But, to date, it has not been reported whether the AAC-11 gene in cervical cancer cells enhances metastatic potential and inhibits apoptosis after growth factor deprivation. Because the CC231 clone was identified only in metastatic tissue and cancer cell line by the mRNA DD method, we have compared the different expression levels of AAC-11 mRNAs in normal cervical, cervical cancer, metastatic tissues, and cervical cancer cell lines, and evaluated whether AAC-11 gene shows metastasis-related and apoptosis-inhibiting functions in cervical cancer cells.
Results
Differential Display and Library Screening
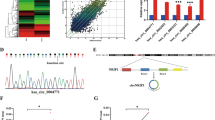
We identified a cDNA PCR fragment, CC231, that was expressed in metastatic tissue and cervical cancer cell line, but not in normal cervical and cervical cancer tissues, using a 5′arbitrary primer H-AP23 (5′-AAGCTTGGCTATG-3′) and a 3′H-T11C anchored primer (5′-AAGCTTTTTTTTTTTC-3′). Searches of NIH GenBank using BLAST programs indicated that the sequence of 491 bp CC231 cDNA PCR product has 98% homology to the sequence at the 3′non-coding region of human AAC-11 gene (accession number U83857). The CC231 cDNA begins at nucleotide 3194 of human AAC-11. To identify potential AAC-11 isoforms, cDNA library constructed from metastatic cancer tissue was screened by Southern hybridization with the random primer labeled fragment of CC231. We isolated a 3685 bp cDNA (data not shown). AAUAAA polyadenylation signal was found in this cDNA. This longer cDNA contains an additional 35 bp in the 3′ end of the original human AAC-11 cDNA.
AAC-11 Gene Expressions
Different expression levels of AAC-11 gene in normal cervical, cervical cancer, metastatic tissues, and cervical cancer cell lines are shown in Figure 1. Northern blot analysis revealed that a dominant transcript of approximately 4.1 kb was present in all tissues and cell lines examined, as well as a weaker band at 1.7 kb. In addition, a transcript of approximately 3.1 kb was identified in all five cell lines tested. The level of expression of AAC-11 mRNA in nine normal cervical tissues (Fig. 1A) and nine primary cervical cancer tissues (Fig. 1B) was low. On the contrary, its expression was higher in three metastatic common iliac lymph node tissues (Fig. 1C). In metastatic tissues, the levels of AAC-11 gene expression were increased by 162 ± 62% as compared with those of gene expressions in cervical cancer tissues. AAC-11 gene expression was found to be expressed in all five cervical cancer cell lines (HeLa, CaSki, SiHa, CUMC-3, and CUMC-6) (Fig. 1D). Among five cancer cell lines, AAC-11 was found to be expressed in high levels in HeLa and CaSki cells when compared with cervical cancer tissues.
Northern blot analysis of AAC-11 mRNA in nine normal cervical tissues, A; nine primary cervical cancer tissues, B; three metastatic lymph node tissues, C; and five cervical cancer cell lines, D. Total RNAs were subjected to electrophoresis through 1% formaldehyde agarose gel and transferred to nylon membranes. The blots were hybridized with 32P-labeled random-primed AAC-11 cDNA probe. A full human β-actin cDNA probe was used as an internal standard to check whether equal amounts of total RNAs were applied.
AAC-11 Gene Transfer and Expression in Cervical Cancer Cells
Human AAC-11 gene was introduced into CUMC-6 cells by DOTAP and the transfected cells were selected by culturing in complete medium containing 0.6 mg/ml of G418 for 21 days. The selected cells were cloned and designated as Clone 1 and Clone 2, respectively. The isolated clones were screened for the overexpression of AAC-11 gene. Northern blot analysis showed that approximately 4.1 kb mRNA single transcript, which was identical to those of the AAC-11 found in metastatic tissues and cervical cancer cell lines, was overexpressed in human AAC-11-transfected Clone 1 and Clone 2, respectively, compared with cells transfected with vector alone or wild-type cells (Fig. 2).
Invasive and Adhesive Properties of AAC-11 Gene
Invasion of matrigel by CUMC-6 cells was increased approximately two times higher in AAC-11-transfected cells (Clone 1) than in cells transfected with vector alone or wild-type cells (Fig. 3A). In Clone 2, invasiveness was increased by approximately 150% compared with vector alone or wild-type cells. Adhesion of CUMC-6 cells transfected with AAC-11 expression vector to laminin was increased by approximately 4-fold in Clone 1 when compared with cells transfected with vector alone or wild-type cells (Fig. 3B). But adhesiveness was increased by approximately 3-fold in Clone 2. Accordingly, we chose Clone 1 for further biological studies of AAC-11.
Comparisons between the invasion and adhesion activities of cervical cancer cells transfected with AAC-11 expression vector, pCR3-Uni vector alone, or wild-type cells. AAC-11 increases invasiveness and adhesiveness in cervical cancer cells. A, In vitro invasiveness of cervical carcinoma cells (Clone 1 and Clone 2) transfected with AAC-11 expression vector, vector alone, or wild-type cells. Invasion assay was carried out as previously described. Five fields were counted for each assay. Each value is the mean ± sd of triplicate determinations. B, Adhesion of cervical carcinoma cells (Clone 1 and Clone 2) transfected with AAC-11 expression vector, vector alone, or wild-type cells to laminin. Adhesion is expressed as the absorbance recorded. Each value is the mean ± sd of quadruplicate determinations.
MMP-2, MT1-MMP, and TIMP-2 Expressions
To evaluate whether MMP-2, MT1-MMP, or TIMP-2 transcript is involved in AAC-11-induced cervical cancer invasion and adhesion, PCR amplifications of each gene were conducted using first-strand cDNAs from CUMC-6 cells transfected with AAC-11 expression vector, cells transfected with vector alone, and wild-type cells, respectively. Utilizing this assay, the predicted 399 bp, 642 bp, and 338 bp size cDNA products of MMP-2, MT1-MMP, and TIMP-2, respectively, were detected in AAC-11 transfected Clone 1 cells (Fig. 4A). But expressions of MMP-2 and MT1-MMP were not detected and expression of TIMP-1 transcript was high in wild-type cells or cells transfected with vector alone, respectively (Fig. 4A). Northern blot analyses revealed that approximately 3.4 kb MMP-2 and 4.1 kb MT1-MMP transcripts were overexpressed in AAC-11 transfected cells (Fig. 4B). In wild-type cells or cells transfected with vector alone, both MMP-2 and MT1-MMP transcripts were not present, respectively. On the contrary, approximately 3.9 kb TIMP-1 transcript was overexpressed in wild-type cells or cells transfected with vector alone. Its expression was undetectable in AAC-11 transfected cells. Cells transfected with AAC-11 expression vector induced increased synthesis of active form of MMP-2 protein, measured by an increase in cellular level of 64-kDa MMP-2 on Western blot. Its expression was undetectable in cells transfected with vector alone (Fig. 4C).
RT-PCR and Northern blot analyses of MMP-2, MT1-MMP, and TIMP-2 gene expressions. Lane 1, wild-type cells; Lane 2, cells transfected with vector alone; and Lane 3, AAC-11-transfected Clone 1 cells. A, PCR amplification of each gene using first-strand cDNAs. The predicted fragments amplified by PCR for MMP-2, MT1-MMP, TIMP-2, and β-actin transcripts are 399, 642, 338, and 350 bp, respectively. Lane M denotes molecular weight size marker. B, Northern blot analyses of transcripts of MMP-2, MT1-MMP, and TIMP-2. cDNA probes were prepared by PCR with digoxigenine-11–2′-deoxy-uridine-5′-triphosphate using corresponding RT-PCR products, respectively. The Northern blot results were normalized with β-actin. C, Increased MMP-2 protein level after AAC-11 gene transfection. Immunoblot analysis of the cell lysates from AAC-11 transfected cells and cells transfected with vector alone were done using an anti-MMP monoclonal antibody. Total cellular proteins from 5 × 105 cells were subjected to SDS-PAGE. Bands were revealed by ECL-Western blot detection kit.
β-catenin Gene Expression
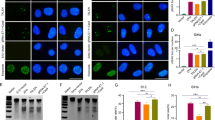
In CUMC-6 cancer cells transfected with AAC-11, the expression of β-catenin was up-regulated as compared with gene expressions in wild-type cells or cells transfected with vector alone, respectively (Fig. 5).
β-catenin gene expression in AAC-11 transfected cervical cancer cells. Total RNAs from wild-type cells, cells transfected with vector alone, and AAC-11 transfected cervical cancer cells were subjected to electrophoresis and transferred to nylon membrane. The blot was hybridized with 32P-labeled random-primed RT-PCR product.
Growth Characteristics
The growth phenotype of AAC-11-transfected Clone 1 cells was assessed by colony-forming assay. AAC-11 gene transfection increased cervical cancer cell colonization up to approximately 140% when compared with cells transfected with vector alone (Fig. 6).
AAC-11 Inhibits Apoptosis
The survival advantage in serum-free medium offered by transfection with AAC-11 is shown in Fig 7. The effect of AAC-11 on cultured cervical cancer cells was associated with antiapoptotic process. Approximately 50% of the AAC-11-transfected Clone 1 cells died after 2 weeks, compared to 1 week for vector alone or wild-type cells. A near-complete loss of viability generally occurred by Days 24 to 28 in both wild-type cells and cells transfected with vector alone. Dying cells showed apoptotic body formations, including fragmented nuclei and chromatin condensation (not shown). On the contrary, AAC-11-transfected cells had a marked survival advantage. Viable transfectants persisted for up to 8 weeks in the absence of serum, and some clones regrew to confluence on the addition of serum.
Discussion
The data presented here indicate that the levels of the AAC-11 gene were increased in metastatic lymph node tissues and cancer cell lines compared with the levels observed in normal cervical and primary cervical cancer tissues. According to this result, we investigated whether the AAC-11 gene is associated with metastatic potential. Our in vitro study using adhesion and invasion assays suggested that AAC-11 may be involved in human cervical cancer cell invasion, probably through its role in cell interactions with extracellular matrix proteins.
MMP-2 (gelatinase A) is able to degrade several extracellular matrix proteins including denatured collagen, type iv collagen, and fibronectin (Liotta et al, 1980). A number of reports have shown that increased levels of MMP-2 are correlated with an invasive phenotype (Tryggvason et al, 1993). MMP-2 can bind and act as a receptor for surface-localized metalloproteinase activity (Brooks et al, 1996), but little is known about cell surface interactions and MMP-2. MT1-MMP is localized on the cell surface, functions as a cell surface receptor in association with TIMP-2, and mediate the activation of proMMP-2 (progelatinase A) (Sato et al, 1994).
MT1-MMP is most often overexpressed on the surface of invasive tumor cells, including lung and stomach cancers that contain activated MMP-2. TIMP-2 has been shown to suppress tumor growth and metastatic potential in many cell model systems, activities associated with inhibition of MMP (Stetler-Stevenson et al, 1990). Recently, MMP-2, MT1-MMP, and TIMP-2 have been investigated regarding the invasion and metastatic potentials of cancer cells (Martorana et al, 1998; Valente et al, 1998).
To confirm whether AAC-11 gene transfection augmented the invasive ability of cancer cells, the levels of MMP-2, MP1-MMP, and TIMP-2 expressions were examined. ACC-11-transfected cervical cancer cells expressed high levels of MMP-2 and MT1-MMP mRNAs. TIMP-2 gene was found to be expressed in high levels only in cells transfected with vector alone or wild-type cells, respectively. These data thus suggest that AAC-11 gene up-regulated the level of MMP-2 and MT1-MMP mRNAs and down-regulated TIMP-1 during the process of cervical cancer invasion and adhesion.
MMP-2 is secreted as proenzymes that require extracellular activation. In vitro studies of the activation mechanism of this enzyme has yielded new insights into the molecular basis of proenzyme latency (Stetler-Stevenson et al, 1991). In this study, AAC-11 transfected cancer cells expressed an elevated level of MMP-2 protein as assessed by immunoblotting. Elevated levels of 64 kDa MMP-2 protein in AAC-11 transfected cervical cancer cells mean that AAC-11 converted the latent form of 72-kDa proMMP-2 to the active form. This finding shows that the AAC-11 gene is capable of regulating the synthesis of MMP-2.
Frequent alterations in β-catenin expression have been reported in various types of human cancers (Takayama et al, 1996). Evidence is now accumulating that deregulated signal transduction involving β-catenin is a common event in tumor cells. β-catenin and its homolog Armadillo are key components of cell-cell adhesive junctions (Peifer, 1997). In this study, the expression of β-catenin was up-regulated in AAC-11 transfected cervical cancer cells. From the above results, we can estimate that overexpression of MMP-2 and MT1-MMP, loss of TIMP-2 expression, and up-regulation of β-catenin by AAC-11 transfection may contribute to the development of cervical cancer invasion.
Tewari et al (1997) identified a 1023-bp AAC-11short cDNA from murine fibroblasts by a functional expression cloning approach. The predicted 25 kDa AAC-11 protein contained a leucine zipper motif that is essential for its antiapoptotic activity. The cDNA for human AAC-11 was also identified and exhibited strong homology with mouse AAC-11long and retained the leucine zipper domain. Expression of the AAC-11 gene prevents apoptosis that occurs on deprivation of growth factors. Inactivating mutations of leucines within the leucine zipper domain of AAC-11short cDNA ablated its antiapoptotic function. We also observed the survival advantage after serum deprivation offered by transfection with AAC-11 cDNA in human cervical cancer cells. Because inhibition of apoptosis is thought to be required for neoplastic transformation, study of the AAC-11 gene and its ability to inhibit apoptosis will provide unique insights into the regulation of cell death.
Further characterization of the AAC-11 gene in regard to its metastasis-related and antiapoptotic functions may lead to a better understanding of the tumor biology of human cervical cancer.
Materials and Methods
Tissues and Cell Lines
For DD of mRNA, normal cervical tissue specimens were obtained from uterine myoma patients during hysterectomy, and untreated primary cervical cancer and metastatic common iliac lymph node tissues were obtained from cervical cancer patients during radical hysterectomy. The human cervical cancer cell line used in the DD method was CUMC-6 (Kim et al, 1996). The human cervical cancer cell lines used in this study were HeLa, CaSki, SiHa, CUMC-3 (Kim et al, 1995), and CUMC-6.
Isolation of Total RNA and DD
Total RNAs were extracted from fresh tissue specimens and cultured cells using a commercial system (RNeasy total RNA kit) provided by Qiagen (Qiagen Inc., Hilden, Germany) and removal of DNA contamination from the RNAs was accomplished using Message clean kit (GenHunter Corp., Brookline, Massachusetts). DD was performed using reverse transcription-polymerase chain reaction (RT-PCR) as described (Liang and Pardee, 1992) with minor modifications. Briefly, triplicate 0.2 μg of total RNA was used for RT with one of three primers, H-T11G, H-T11C, or H-T11A, as the anchored oligo-dT primers (RNAimage kit; GenHunter), followed by PCR using the same anchored primers and one of the arbitrary 5′ 13 mer (RNAimage primer sets 3 and 4, H-AP 17–32) in the presence of 0.5 mmol/L of [a-35S]-labeled dATP (1200 Ci/mmol). PCR was 40 cycles at 95° C for 40 seconds, 40° C for 2 minutes and 72° C for 40 seconds, followed by 72° C for 5 minutes. PCR-amplified fragments were resolved in 6% polyacrylamide sequencing gels. The gels were dried under vacuum on a gel dryer at 80° C for 1 hour. Differentially expressed fragments were identified by inspection of autoradiograms.
Band Recovery and Characterization
Bands of interest were excised from the dried sequencing gel. The cDNAs were eluted by boiling the gel slices for 15 minutes and were reamplified with the same primer pairs. PCR conditions were the same as those for amplification, except that no [a-35S]-labeled dATP and 20 μmol/L of dNTPs were used. The reamplified cDNAs were cloned into the pGEM-T Easy vector using the TA Cloning System (Promega, Madison, Wisconsin), following the manufacturer's instructions. The clones that had inserts with the size of the original cDNA were sequenced using the Sequenase Version 2.0 DNA Sequencing System (United States Biochemical Co., Cleveland, Ohio).
Construction of cDNA Library and Library Screening
To isolate potential AAC-11 isoforms, a cDNA library was constructed from 1 μg of oligodeoxythymidine-selected RNA using a SMART PCR cDNA library construction kit (Clontech, Palo Alto, California) according to the procedure recommended by the manufacturer. Oligodeoxythymidine-selected RNA from metastatic cervical cancer tissue was used for the construction of the cDNA library. The cDNA library was screened by Southern hybridization of duplicate filter lifts (1 × 108 pfu) with the random primer labeled fragment of CC231 (Feinberg and Vogelstein, 1983; Sambrook et al, 1989).
Construction of Recombinant Plasmid Expressing Human AAC-11 cDNA
Single-stranded AAC-11 cDNA was synthesized using a total RNA from primary cervical cancer tissue. Deoxyoligonucleotide primers were designed according to the published sequences from untranslated regions of AAC-11 mRNA (accession number U83857) and were used in PCR to amplify AAC-11 cDNA. The primer sequences were as follows: the forward primer, 5′-GGCGCCGGTCAGGACAAGGATA-3′(positions 43–64) and the reverse primer, 5′-TCCCCGGCAATCCCAA- TGAGTAGT-3′(positions 1720–1743). The amplified 1701 bp AAC-11 cDNA fragment was ligated into the expression vector pCR3-Uni (InVitrogen, San Diego, California). The specificity of the amplified AAC-11 PCR product was verified with DNA sequencing.
Northern Blot Analysis of AAC-11 Gene
To determine the level of AAC-11 gene expression, 20 μg of denatured total RNAs were subjected to electrophoresis through 1% formaldehyde agarose gel and transferred to nylon membranes (Boehringer-Mannheim Biochemica, Mannheim, Germany). The blots were hybridized with 32P-labeled random-primed 491 bp AAC-11 cDNA probe overnight at 42°C. The northern blot results were consistently repeated two times, as quantified by densitometry and normalized with β-actin.
Transfection of AAC-11 Gene into Cervical Cancer Cells
Synthetic cationic liposome, DOTAP {N-[1-(2, 3-Dioleoyloxy)propyl]–N, N,N-trimethylammonium-methylsulfate} (Boehringer Mannheim), was used to introduce the AAC-11 expression vector into CUMC-6 cells (Holmen et al, 1995). Briefly, 5 × 105 cells were seeded in 60-mm tissue culture dishes (Costar, Cambridge, Massachusetts). After overnight incubation in a humidified 5% CO2 incubator, the cells were treated with 150 μl of DOTAP-DNA complex containing 30 μg of DOTAP reagent and 5 μg of CsCl gradient banded AAC-11 DNA. After 16 hours of incubation, the cells were split at a ratio of 1:5 and cultured on complete medium containing G418 (Gibco BRL, Gaithersburg, Maryland). Cells were selected for resistance to 0.6 mg/ml of G418 for 21 days.
Selected transfectants were cloned at 0.5 cell/well, and maintained in medium containing 0.6 mg/ml of G418. Two G418-resistant colonies were isolated and designated as Clone 1 and Clone 2, respectively. Another population of CUMC-6 cells was transfected with expression vector pCR3-Uni alone and similarly selected for G418 resistance to serve as an experimental control.
Invasion Assay
In vitro invasiveness was carried out by the method of Albini et al, (1987) with minor modifications. Invasion was measured by use of 24-well transwell units with 12-μm porosity polycarbonate filters coated with the reconstituted basement membrane substance (Matrigel; Collaborative Research, Lexington, Massachusetts). The filters in the transwells were coated with 30 μg of matrigel per filter in cold serum-free medium to form a thin, continuous layer on the top of the filter. The matrigel was left to air-dry overnight. Before addition of the cells, excess medium was removed from the upper compartment. The lower compartment contained 0.5 ml of serum-free medium supplemented with 10 ng/ml of platelet-derived growth factor (Sigma) as a chemoattractant. Cells were then harvested by trypsinization and counted; then 5 × 104 cells were resuspended in 100 μl of serum-free medium and placed in the upper compartment of the transwell unit for 5 hours at 37° C. Cells were fixed with methanol and stained with Wright. Cells on the upper surface of the filter were removed by wiping with a cotton swab, and invasion was determined by counting the cells that had migrated to the lower side of the filter, using a light microscope at ×200 magnification. Five fields were counted for each assay. Each sample was assayed in triplicate.
Adhesion Assay
The adhesion assay was carried out by the method of Merzak et al (1994) with minor modifications. Briefly, 96-well plates were coated overnight with laminin (Sigma) at 10 μg/ml. Cells were then harvested by trypsinization and counted; then 5 × 104 cells were resuspended in 200 μl of serum-free medium and placed in the laminin-coated plates. After 7 hours of incubation, the attachment was measured by using the MTT assay kit (Chemicon International, Temecula, California). During the last 4 hours of culture, 10 μl of MTT in PBS was added to each well. At the end of this time, 100 μl of isopropanol/0.04 N HCl was added and the absorbency was measured on an ELISA plate reader (Dynatech MR 700 reader; Dynatech Labs, Chantilly, Vermont) with a test wavelength of 570 nm. All samples were plated in quadruplicate. Cell attachment was expressed as the absorbency recorded.
RT-PCR and Northern Blot Analyses of the MMP-2, MT1-MMP and TIMP-2 Genes
Total RNAs from cervical cancer cells transfected with AAC-11 gene, expression vector pCR3-Uni alone or wild-type parental cells were transcribed using random hexamer (Perkin-Elmer Cetus, Foster City, California) and Moloney murine leukemia reverse transcriptase (Promega), respectively. Twenty-μl of the cDNA reaction mixture was incubated at 42° C for 60 minutes, heated at 95° C for 5 minutes, and then slowly cooled to 4° C. PCR was carried out in a 20-μl reaction containing 1.5 μl of RT reaction mixture, 100 μmol/L of dNTPs, 10 pmol each of oligonucleotide primer, 10 mmol/L of KCl, 10 mmol/L of Tris-HCl (pH 8.3), 2.5 mmol/L of MgCl2, and 0.5 units of Taq polymerase. The PCR mixture was overlaid with mineral oil and then amplified in a thermal cycler (Perkin-Elmer) for 35 cycles. The amplification profile for MMP-2 and MT1-MMP was as follows: denaturation at 94° C for 30 seconds, primer annealing at 60° C for 45 seconds, and extension at 72° C for 15 seconds. For TIMP-2 and β-actin amplifications, the temperature for primer annealing was reduced to 58° C. The sequences of oligonucleotide primers, synthesized according to the published MMP-2 (accession number J03210), MT1-MMP (accession number X90925), TIMP-2 (accession number J05593), and β-actin cDNA (Ng et al, 1985) sequences, were as follows: for the MMP-2, forward, 5′-ATGACAGCTGCACCACT- GAG-3′ and reverse, 5′-GGGACAGACGGAAGTT- CTTG; for the MT1-MMP, forward, 5′-AACATTGGA- GGAGACA-CCCA-3′ and reverse, 5′-CCAGAAGA- GAGCAGCATCAA-3′; for the TIMP-2, forward, 5′-AGCA-CCACCCAGAAGAAGAG-3′ and reverse, 5′-CCAGGAAGGGATGT-CAGAG-3′; for the β-actin, forward, 5′-AGGCCAACCGCGAGAAG-ATGACC-3′ and reverse, 5′-GAAGTCCAGGGCGACGTAGCAC-3′.
Amplified products were subjected to electrophoresis through 1.2% agarose gels and visualized by ethidium bromide staining. The predicted fragments amplified by PCR for MMP-2, MT1-MMP, TIMP-2, and β-actin transcripts were 399, 642, 338, and 350 bp, respectively.
For Northern blot analyses, MMP-2, MT1-MMP, TIMP-2, and β-actin cDNA probes were prepared by PCR with digoxigenine-11–2′-deoxy-uridine-5′-triphosphate (Boehringer-Mannheim) using corresponding RT-PCR products. The blots were washed and incubated with alkaline phosphatase-conjugated anti-digoxigenine antibody and then hybridized probes were immunodetected with chemiluminescent substrate supplied by Boehringer-Mannheim.
Western Blot Analysis
For immunoblot analysis, 5 × 105 cells were harvested and lysed in Laemmli sample buffer (Laemmli, 1970). The cellular proteins were separated by 10% sodium dodecylsulfate polyacrylamide gel electrophoresis (SDS-PAGE) and then electroblotted onto nitrocellulose membranes.
After blocking with 5% non-fat milk in PBS, the membranes were incubated with an anti-human mouse MMP-2 antibody (R and D Systems, Minneapolis, Minnesota) for 16 hours at 4° C. With being washed by PBS containing 0.2% Tween three times per 15 minutes, membranes were incubated with the blocking solution containing 1:1000 dilution of horseradish peroxidase-conjugated goat anti-mouse immunoglobulin (Amersham) as a secondary antibody. Proteins were revealed by ECL-Western blot detection kit (Amersham).
Expression of β-catenin Gene
Total RNA from normal cervical tissue was transcribed using random hexamer (Perkin-Elmer) and Moloney murine leukemia reverse transcriptase (Promega). The amplification profile for β-catenin was as follows: denaturation at 94°C for 30 seconds, primer annealing at 60°C for 45 seconds, and extension at 72°C for 15 seconds. The sequences of oligonucleotide primers, synthesized according to the published β-catenin (accession number X87838) sequence, was as follows: for the β-catenin, forward, 5′-TCGGGCTGGTGAC- AGGGAAGACAT-3′ and reverse, 5′-CATGGACG- TACGGCGCTGGGTATC-3. The predicted fragment amplified by PCR for β-catenin transcript was 304 bp (not shown). To determine the level of β-catenin expression, 20 μg of denatured total RNAs were subjected to electrophoresis and transferred to nylon membranes. The blots were hybridized with 32P-labeled random-primed RT-PCR product overnight at 42° C.
Growth Characteristics
AAC-11 gene transfected cells were studied to estimate the colony-forming efficiency. To determine the colony-forming efficiency, 5 × 103 viable cells were suspended in 1 ml of 0.3% noble agar (Difco Laboratories Inc., Detroit, Michigan) made with complete media, and layered onto 0.6% agar in 35-mm plates. All samples were plated in triplicate. The number of CUMC-6 cell colonies (>50 cells/cluster) was estimated on Day 21.
Survival Assay
To test the effect of AAC-11 on cervical cancer cell survival, 5 × 105 wild-type CUMC-6 cells, AAC-11 gene transfected cells, and cells transfected with pCR3-Uni alone were cultured in serum-free medium for up to 8 weeks. The survival advantage offered by transfection with AAC-11 gene was observed. In three independent experiments, cells in triplicate flasks were detached and viable cells counted twice a week using trypan blue dye exclusion.
References
Albini A, Iwamoto Y, Kleinman HK, Martin GR, Aarronson SA, Kozlowski JM, and McEwan RN (1987). A rapid in vitro assay for quantitating the invasive potential of tumor cells. Cancer Res 47: 3239–3245.
Brooks PC, Stromblad S, Sanders LC, von Schalscha TL, Aimes RT, Stetler-Stevenson WG, Quigley JP, and Cheresh DA (1996). Localization of matrix metalloproteinase MMP-2 to the surface of invasive cells by interaction with integrin αvβ3. Cell 85: 683–693.
Choo KB, Chong KY, Chou HF, Liew LN, and Liou CC (1989). Analysis of the structure and expression of the c-myc oncogene in cervical tumor and in cervical tumor-derived cell lines. Biochem Biophys Res Commun 158: 334–340.
Feinberg AP and Vogelstein B (1983). A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem 132: 6–13.
Fujita M, Inoue M, Tanizawa O, Iwamoto S, and Enomoto T (1992). Alterations of the p53 gene in human primary cervical carcinoma with and without human papillomavirus infection. Cancer Res 52: 5323–5328.
Holmen SL, Vanbrockin MW, Eversole RR, Stapleton SR, and Ginsberg LC (1995). Efficient lipid-mediated transfection of DNA into primary rat hepatocytes. In Vitro Cell Dev Biol Anim 31: 347–351.
Kim JW, Cho YH, Lyu MS, Lee CG, Jung JK, Kim SJ, and Namkoong SE (1995). Establishment and characterization of a cell line (CUMC-3) derived from a human squamous carcinoma of the uterine cervix. Gynecol Oncol 57: 47–60.
Kim JW, Lee CG, Cho YH, Kim JH, Kim SJ, Kim HK, Park TC, Song SK, and Namkoong SE (1996). CUMC-6, a new diploid human cell line derived from a squamous carcinoma of the uterine cervix. Gynecol Oncol 62: 230–240.
Kristensen GB, Holm R, Abeler VM, and Trop CG (1996). Evaluation of the prognostic significance of cathepsin D, epidermal growth factor receptor, and c-erbB-2 in early cervical squamous cell carcinoma: An immunohistochemical study. Cancer 78: 433–440.
Laemmli UK (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685.
Liang P and Pardee AB (1992). Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science 257: 967–971.
Liotta LA, Tryggvason K, Garbisa S, Ian H, Foltz CM, and Shafie S (1980). Metastatic potential correlates with enzymatic degradation of basement membrane collagen. Nature 284: 67–68.
Martorana AM, Zheng G, Crowe TC, O'Grady RL, and Lyons JG (1998). Epithelial cells up-regulate matrix metalloproteinases in cells within the same mammary carcinoma that have undergone an epithelial-mesenchymal transition. Cancer Res 58: 4970–4979.
Merzak A, Koocheckpour S, and Pilkington GJ (1994). CD44 mediates human glioma cell adhesion and invasion in vitro. Cancer Res 54: 3988–3992.
Ng SY, Gunning P, Eddy R, Ponte P, Leavitt J, Shows T, and Kedes L (1985). Evolution of the functional human beta-actin gene and its multi-pseudogene family: conservation of noncoding regions and chromosomal dispersion of pseudogenes. Mol Cell Biol 5: 2720–2732.
Park TW, Fujiwara H, and Wright TC (1995). Molecular biology of cervical cancer and its precursors. Cancer 76: 1902–1913.
Peifer M (1997). Beta-catenin as oncogene: the smoking gun. Science 275: 1752–1753.
Sambrook J, Fritsch EF, and Maniatis T (1989). Molecular cloning: A laboratory manual. New York: Cold Spring Harbor Laboratory.
Sato H, Takino T, Okada Y, Cao J, Shinagawa A, Yamamoto E, and Seiki M (1994). A matrix metalloproteinase expressed on the surface of invasive tumor cells. Nature 370: 61–65.
Stetler-Stevenson WG, Brown PD, Onisto M, Levy AT, and Liotta LA (1990). Tissue inhibitor of metalloproteinase-2 (TIMP-2) mRNA expression in tumor cell lines and human tumor tissues. J Biol Chem 23: 13933–13938.
Stetler-Stevenson WG, Talano JA, Gallager ME, Krutzsch HC, and Liotta LA (1991). Inhibition of human type IV collagenase by a highly conserved peptide sequence derived from its prosegment. Am J Med Sci 302: 163–170.
Takayama T, Shiozaki H, Shibamoto S, Oka H, Kimura Y, Tamura S, Inoue M, Monden T, Ito F, and Monden M (1996). Beta-catenin expression in human cancers. Am J Pathol 148: 39–46.
Tewari M, Yu M, Ross B, Dean C, Giordano A, and Rubin R (1997). AAC-11, a novel cDNA that inhibits apoptosis after growth factor withdrawal. Cancer Res 57: 4063–4069.
Tryggvason K, Hoyhtya M, and Pyke C (1993). Type IV collagenase in invasive tumors. Breast Cancer Res Treat 24: 209–218.
Valente P, Fassina G, Melchiori A, Masiello L, Cilli M, Vacca A, Onisto M, Santi L, Stetler-Stevenson WG, and Albini A (1998). TIMP-2 overexpression reduces invasion and angiogenesis and protects B16F10 melanoma cells from apoptosis. Int J Cancer 75: 246–253.
Wright TC, Ferenczy AF, and Kurman RJ (1994). Precancerous lesions of the cervix. In: Kurman RJ, editor. Blaustein's pathology of the female genital tract. New York: Springer-Verlag, 229–278.
zur Hausen H (1991). Viruses in human cancer. Science 254: 1167–1173.
Author information
Authors and Affiliations
Corresponding author
Additional information
This work was supported by a grant of the 1999 Good Health R&D Project, Ministry of Health and Welfare, Republic of Korea (#HMP-99-B-02–0002).
Rights and permissions
About this article
Cite this article
Kim, J., Cho, H., Kim, J. et al. AAC-11 Overexpression Induces Invasion and Protects Cervical Cancer Cells from Apoptosis. Lab Invest 80, 587–594 (2000). https://doi.org/10.1038/labinvest.3780063
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.3780063
This article is cited by
-
Altered expression of anti-apoptotic protein Api5 affects breast tumorigenesis
BMC Cancer (2023)
-
API5 confers cancer stem cell-like properties through the FGF2-NANOG axis
Oncogenesis (2017)
-
Nucleoprotein of influenza A virus negatively impacts antiapoptotic protein API5 to enhance E2F1-dependent apoptosis and virus replication
Cell Death & Disease (2015)
-
Apoptosis inhibitor-5 overexpression is associated with tumor progression and poor prognosis in patients with cervical cancer
BMC Cancer (2014)