Abstract
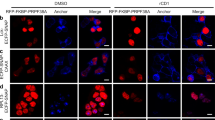
IN mammalian nuclei, precursor messenger RNA splicing factors are distributed non-uniformly. Antibodies directed against struc-tural polypeptides of small nuclear ribonucleoprotein particles (snRNPs)1 and some non-snRNP splicing factors2 have shown that these components are concentrated in about 20-50 nuclear 'speckles'. These and other non-homogeneous distributions have been proposed to indicate nuclear 'compartments' that are distinct from the sites of transcription and in which RNA processing occurs3–9. We have tested this idea using a new approach. Previous structural10–13 and biochemical14–16 data have shown that splicing can occur in association with transcription. Nascent RNA of specific genes can be detected by in situ hybridization as intense spots of nuclear stain which map to the sites of transcription17–19. Here we identify active pre-mRNA splicing sites by localizing the nascent spliced mRNA of specific genes. We find that splicing occurs at the sites of transcription, which are not coincident with intranuclear speckles. We conclude that the nucleus is not compart-mentalized with respect to transcription and pre-mRNA splicing.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Spector, D. L. Proc. natn. Acad. Sci. U.S.A. 87, 147–151 (1990).
Fu, X. D. & Maniatis, T. Nature 343, 437–441 (1990).
Spector, D. L. A. Rev. Cell Blol. 9, 265–315 (1993).
Xing, Y. & Lawrence, J. B. Trends Cell Biol. 3, 346–353 (1993).
Rosbash, M. & Singer, R. H. Cell 75, 399–401 (1993).
Carter, K. C., Taneja, K. L. & Lawrence, J. B. J. Cell Biol. 115, 1191–1202 (1991).
Spector, D. L., Fu, X. D. & Maniatis, T. EMBO J. 10, 3467–3481 (1991).
Huang, S. & Spector, D. L. Genes Dev. 5, 2288–2302 (1991).
Xing, Y., Johnson, C. V., Dobner, P. R. & Lawrence, J. B. Science 259, 1326–1330 (1993).
Sass, H. & Pederson, T. J. molec. Biol. 180, 911–926 (1984).
Beyer, A. L. & Osheim, Y. N. Genes Dev. 2, 754–765 (1988).
Vazquez-Nin, G. H., Echeverria, O. M., Fakan, S., Leser, G. & Martin, T. E. Chromosoma 99, 44–51 (1990).
Wu, Z., Murphy, C., Callan, H. G. & Gall, J. G. J. Cell Biol. 113, 465–483 (1991).
Aebi, M. & Weissman, C. Trends Genet. 3, 102–107 (1987).
LeMaire, M. F. & Thummel, C. S. Molec. cell. Biol. 10, 6059–6063 (1990).
Baurén, G. & Wieslander, L. Cell 76, 183–192 (1994).
O'Farrell, P. H., Edgar, B. A., Lakich, D. & Lehner, C. F. Science 246, 635–640 (1989).
Shermoen, A. W. & O'Farrell, P. H. Cell 67, 303–310 (1991).
Lawrence, J. B., Singer, R. H. & Marselle, L M. Cell 57, 493–502 (1989).
Valcarcel, J., Singh, R., Zamore, P. D. & Green, M. R. Nature 362, 171–175 (1993).
Turner, B. M. & Franchi, L. J. Cell Sci. 87, 269–282 (1987).
Voelkerding, K. & Klessig, D. F. J. Virol. 60, 353–362 (1986).
Bridge, E., Carmo-Fonseca, M., Lamond, A. I. & Pettersson, U. J. Virol. 67, 5792–5802 (1993).
Wansink, D. G. et al. J. Cell Biol. 122, 283–293 (1993).
Jackson, D. A., Hassan, A. B., Errington, R. J. & Cook, P. R. EMBO J. 12, 1059–1065 (1993).
Fakan, S., Leser, G. & Martin, T. E. J. Cell Biol. 98, 358–363 (1984).
Jiménez-García, L. F. & Spector, D. L. Cell 73, 47–59 (1993).
Zachar, Z., Kramer, J., Mims, I. P. & Bingham, P. M. J. Cell Biol. 121, 729–742 (1993).
Séraphin, B. & Rosbash, M. Cell 59, 349–358 (1989).
Taneja, K. L., Lifshitz, L. M., Fay, F. S. & Singer, R. H. J. Cell Biol. 119, 1245–1260 (1992).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Zhang, G., Taneja, K., Singer, R. et al. Localization of pre-mRNA splicing in mammalian nuclei. Nature 372, 809–812 (1994). https://doi.org/10.1038/372809a0
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/372809a0
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.