Abstract
Hepatocyte paraffin 1 (Hep Par 1), a murine monoclonal antibody, is widely used in surgical pathology practice to determine the hepatocellular origin of neoplasms. However, identity of the antigen for Hep Par 1 is unknown. The aim of this study was to characterize the Hep Par 1 antigen. To identify the antigen, immunoprecipitation was used to isolate the protein from human liver tissue, and a distinct protein band was detected at approximately 165 kDa. The protein band was also present in small intestinal tissue, but was not present in several other non-liver tissues nor in three human hepatocellular carcinoma cell lines, Huh-7, HepG2, and LH86. The protein was purified and analyzed by mass spectrometry. It was identified as carbamoyl phosphate synthetase 1 (CPS1). CPS1 is a rate-limiting enzyme in urea cycle and is located in mitochondria. We demonstrated that hepatoid tumors (gastric and yolk sac) were immunoreactive with both Hep Par 1 antibody and anti-CPS1 antibody, further confirming the results of mass spectrometric analysis. We found that the three human hepatocellular carcinoma cell lines do not express either CPS1 RNA or protein. We confirmed that the gene was present in these cell lines, suggesting that suppression of CPS1 expression occurs at the transcriptional level. This finding may have relevance to liver carcinogenesis, since poorly differentiated hepatocellular carcinomas exhibit poor to absent immunoreactivity to Hep Par 1. In conclusion, we have identified the antigen for Hep Par 1 antibody as a urea cycle enzyme CPS1. Our results should encourage further investigation of potential role that CPS1 expression plays in liver pathobiology and carcinogenesis.
Similar content being viewed by others
Main
The histological distinction between hepatocellular carcinomas (HCC) and metastatic adenocarcinoma to the liver can sometimes be a challenging dilemma for surgical pathologists, particularly given the histological variants of HCC that can occur. In addition, tumors in other sites can display hepatoid morphologic features, adding to the diagnostic challenge when considering their metastasis to the liver. In the end, a wide panel of immunohistochemical markers is often used for the differential diagnosis of HCC, cholangiocarcinoma and metastatic adenocarcinoma. These markers include alpha-fetoprotein (AFP), polyclonal carcinoembryonic antigen (pCEA), and alpha-1-antitrypsin.1 None of these markers, however, are highly specific or sensitive for hepatocyte differentiation.
In 1993, Wennerberg et al,2 developed a new monoclonal antibody specific for hepatocytes in formalin-fixed, paraffin-embedded tissues. This antibody, named hepatocyte paraffin 1 (Hep Par 1, clone OCH1E5.2.10), was generated using tissue extracts from a formalin-fixed failed allograft liver. Subsequent studies showed a high sensitivity and specificity of Hep Par 1 for normal hepatocytes and neoplastic hepatic tissue.3, 4, 5, 6 Additional recent studies examined Hep Par 1 reactivity in a variety of non-hepatic tissues, both benign and neoplastic.7, 8, 9, 10 A large number of gastric adenocarcinomas show Hep Par 1 reactivity.11 Rare examples of cholangiocarcinomas, yolk sac tumor, and adenocarcinomas of the ovary, adrenal cortex, lung, endocervix, colon, and pancreas have shown focal Hep Par 1 staining.9, 12, 13 Hep Par 1 reactivity is also seen in benign small intestinal mucosa and intestinal metaplasia of the esophagus and stomach.14
Although the Hep Par 1 antibody exhibits excellent sensitivity and specificity for well-differentiated hepatocytes, the identity of the antigen and its role in liver biology and carcinogenesis are still unknown, constituting one of the great enigmas of diagnostic pathology. Establishing the identity of the antigen is critical, for a number of reasons. First, by knowing the identity of this antigen, additional antibodies of potentially greater diagnostic utility may then be generated. Second, this antigen is predominantly liver-specific. Examining expression patterns for this protein in normal and cancerous liver tissues will likely benefit studies on liver cell biology and pathobiology. Third, such identification may be of substantial value in understanding the significance of the Hep Par 1 reactivity in a number of non-liver cancers, particularly since such expression is associated with a ‘hepatoid’ cellular phenotype.
Herein we report the identification and characterization of the Hep Par 1 antigen. Through the use of immunoprecipitation combined with mass spectrometry, this antigen is identified as carbamoyl phosphate synthetase 1 (CPS1), a relatively liver-specific, intramitochondrial rate-limiting enzyme in the urea cycle. Moreover, our data show that CPS1 is not expressed in human liver cancer cell lines in cell culture, raising the question of whether suppression of CPS1 expression has relevance to liver carcinogenesis.
MATERIALS AND METHODS
Tissue Samples
A total of five liver tissue samples, obtained from the University of Florida Shands Hospital, were used to establish the identity of the Hep Par 1 antigen. These tissue samples include normal liver resected for metastatic colon cancer (three patients, two males and female) and primary hepatocellular carcinoma tissue (two patients, males). None of the liver tissues were positive for any known viral infections. Normal non-liver tissue samples from the surgical bench were used to determine the liver specificity of the Hep Par 1 antigen. The normal tissue collected included: skin, thyroid, small intestine, appendix, gallbladder, kidney, and spleen. The surgical pathology case files at the University of Florida from 2000 to 2007 were searched to identify non-liver tumors with ‘hepatoid’ differentiation (cohesive polygonal tumor cells with abundant eosinophilic cytoplasm). One gastric carcinoma was identified with focal ‘hepatoid’ histological features. Three yolk sac tumors also were identified, one of which exhibited focal ‘hepatoid’ differentiation. Paraffin blocks were sectioned for the purposes of CPS1 and Hep Par 1 immunohistochemistry.
A Chinese hamster cell line (CHO) was used as a negative control for immunological reactivity to Hep Par 1. Protein extract also was prepared from three hepatocellular carcinoma cell lines, Huh-7, HepG2, and LH86.15 Tissues and cell line homogenates were homogenized in protein lysis buffer (150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris-CL pH 8.0, 2 mM DTT, 40 μg/ml PMSF, 2 μg/ml aprotinin, 2 μg/ml leupeptin) followed by centrifugation (13 000 r.p.m., 8000 g) at 4°C for 10 min. Protein concentrations were determined from the supernatants using the BioRad Lowry Protein Assay and protein spectrophotometer (750 nm) as previously reported.16
Immunohistochemistry Staining
Immunohistochemistry, using Hep Par 1 antibody (obtained from DAKO, Carpinteria, CA) was performed on frozen liver tissue sections to determine if antigen detection was possible on non-formalin-fixed tissue. Sections (5 μm thick) were obtained in a freezing microtome and fixed in ethanol without any formalin exposure. Alternatively, formalin fixation of tissue, paraffin embedding, and sectioning (5 μm thick) were performed by routine methodology using antigen retrieval. Immunohistochemical reactions with Hep Par 1 were as reported previously.10 The antibody dilution is 1:50. Polyclonal anti-CPS1 antibody was purchased from Abcam (Cambridge, UK).
Immunoprecipitation
Immunoprecipitation was used to purify the Hep Par 1 antigen from the liver tissue samples. For each sample, 60 μl (100 μg protein) of liver protein extract was added to a solution combination of protein lysis buffer as described above, and fresh proteinase inhibitor (40 μg/ml PMSF, 2 mM DTT, 2 μg/ml Aprotinin, 2 μg/ml Leupeptin) to a final volume of 500 μl. The resulting tissue extract solution was incubated with 7.5 μg of Hep Par 1 monoclonal antibody (135 mg/l concentration) overnight on a 4°C rocker. After overnight incubation, 50 μl of protein G agarose/sehparose bead slurry (Sigma, St Louis, MO, USA) was added to the antibody/tissue extract solution and incubated at 4°C for 2 h on a tube rocker. The beads were collected by centrifugation at 13 000 r.p.m. at 4°C for 5 min. The agarose beads were washed three times with 500 μl of cold 1 × TBS buffer with 0.1% Tween 20. The beads were resuspended in 20 μl of 2 × SDS sample buffer, boiled for 5 min, and collected by centrifugation. The resulting supernatant was analyzed by SDS-polyacrylamide gels (SDS-PAGE) as previously reported.17
Western Blot Analysis
Approximately 40 μg of total protein extracts from the liver tissue or non-liver tissues were loaded on 10% SDS-polyacrylamide gels. The gels were run at 100 V for approximately 1 h. The gels were electrophoretically transferred to polyvinylidene difluoride membrane (PVDF) (BIO-RAD, Hercules, CA, USA). The membranes were incubated overnight at 4°C in blocking buffer (1 × TBS containing 0.1% Tween 20 and 5% fat-free milk power (wt/vol)). Hep Par 1 monoclonal antibody (1:500 dilution) was added to the liver extract membrane and the non-liver tissue membranes in 1% bovine serum albumin (BSA) in 1% TBS containing 0.1% Tween 20 for 1 h on a rocker. Anti-actin antibody (1:1000 dilution) was used for an internal control. The membranes were washed three times for 10 min each with TBS containing 0.1% Tween 20, followed by incubation with goat anti-mouse IgG-HRP from Santa Cruz Biotechnology Inc. (Santa Cruz, CA, USA) (dilution 1:2000) in TBS containing 0.1% Tween 2 0 and 5% fat-free milk for 1 h. After washing for three times as described above, the membrane was incubated with Supersignal West Pico Chemilunescent Substrate (Pierce Biotechnology, Rockford, IL, USA) solution for signal detection.
Mass Spectrometry
Gel pieces excised from the SDS-PAGE gels were washed twice with 50 μl of destain solution (50% v/v acetonitrile, 25 mM NH4HCO3) for 45 min to remove the Coomassie blue stain. The pieces were dried for 20 min. In-gel tryptic digestions were carried out by adding 15 μl, 5.0 mM NH4HCO3 containing 12.5 ng/ml trypsin and incubating at 37°C for 24 h. Peptides were extracted by incubation of the gel pieces with 10 μl of 50% v/v acetonitrile in water containing 0.1% of trifluoroacetic acid for 15 min. The extracted tryptic digest peptides were analyzed by liquid chromatography–electrospray tandem mass spectrometry (LC/MSMS). The liquid chromatographic separation of the peptide digests was run on a capillary C18 column with dual mobile phases of a gradient of ACN in water containing 0.1% HAc, and the mass spectrometric identification data were acquired using an LCQ deca mass spectrometer (Thermo Finnigan) performed in positive electrospray and data-dependent mode. The MSMS data for the digest sample from the distinct gel band were searched using MASCOT ion MSMS search against the NCBI_nr database with peptide tolerance of 2.0 Da and MS/MS tolerance of 0.8 Da. One missed cleavage for the trypsin digestion was allowed in the database search.
Cell Culture, RNA Extraction, RT-PCR, and PCR Analysis
All the three hepatocellular cell lines were cultured in DMEM medium with 10% fetal calf serum, as described before.18 Primary human hepatocytes were obtained and cultured according to published protocol.19 Procedures of RNA and DNA extraction from liver cancer cell lines, PCR, and RT-PCR analysis were previously published.16 The primers for CPS1 RT-PCR analysis were: 5′ATGCGGCCGCAGCCACAAATCATCTTCAAAA; 5′ CGTCTAGATGAGAAAGTTGTGAATCAGTTCC, and primers for PCR analysis were 5′-CAACTCAGCATGGCATTGAC; 5′-ACAGCTGTCCTCCGAATCAC.
RESULTS
Hep Par 1 Antibody Detects a 165 kDa Protein in Liver Tissue
Hep Par 1 antibody was originally derived using formalin-fixed liver tissue.2 Immunostains using this antibody in pathology practice are almost exclusively performed on formalin-fixed and paraffin-embedded tissue sections. It is unknown whether the antigen detection needs formalin fixation. Identification of the protein using formalin-fixed tissue would be extremely difficult due to protein crosslinking following formalin exposure. To answer this question, we performed immunohistochemical staining using Hep Par1 antibody on fresh frozen liver tissue sections, post-fixed with ethanol without any formalin exposure. As shown in Figure 1, Hep Par 1 reacted with the antigen in the fresh frozen liver tissue, as demonstrated by granular staining similar to formalin-fixed liver tissue section. This suggests that Hep Par 1 antigen is not formalin fixation or paraffin-induced, although its name connotes paraffin (hence the abbreviation ‘Hep Par 1’). This finding justified use of fresh liver tissue to identify this antigen.
Immunohistochemistry of liver tissue with Hep Par 1 antibody. Normal liver tissue was used for immunoperoxidase staining with the Hep Par 1 antibody (Dako Cat#: M-7158, 1:50 dilution). (a) Negative control for Hep Par 1 immunostain, on fresh frozen liver tissue fixed with ethanol. (b) Hep Par 1 antibody immunostain of fresh frozen liver fixed with ethanol. (c) Hep Par 1 antibody immunostain in formalin-fixed liver tissue. Original magnification × 200.
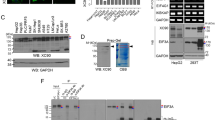
We next prepared protein extracts from fresh normal liver tissue, a human hepatocellular carcinoma cell line, Huh-7 as a potential positive, and the CHO cell line as a presumed negative. The protein extracts were resolved by SDS-PAGE followed by western blot analysis using the Hep Par 1 antibody. As shown in Figure 2, an abundant and specific 165 kDa band is detected only in the fresh liver extracts, but not in the Huh7 or CHO cell lines. This protein was designated as p165. The Huh7 cell line represents a poorly differentiated cancer cell line. Apparently, p165 is not present in this cell line.
Western blot analysis using Hep Par 1 antibody. Protein extracts were made from normal liver tissue, Huh7, and CHO cell lines. Equal amounts of protein extracts were resolved in 10% SDS-PAGE, followed by western blot analysis using Hep Par 1 antibody (1:500 dilution). The antibody detected a 165 kDa band only in the liver tissue extracts (two separate tissue samples in two lanes).
Hep Par 1 Antigen is Present in Liver and Small Intestine Epithelium
The tissue distribution of p165 was next examined using western blot analysis. We prepared protein extracts from normal gallbladder, thyroid, small intestine, spleen, kidney, and skin. These protein extracts were analyzed by western blot analysis with Hep Par 1 antibody. The result revealed a distinct p165 band in the small intestine tissue, but not in other non-liver tissues examined (Figure 3). We then stained a section of normal intestinal tissue using Hep Par 1 antibody. As shown in Figure 3b, strong immunohistochemical reaction is present in the epithelial cells of the small intestine, similar to what has been reported before.20
Immunoreactivity of Hep Par 1 antibody of non-liver normal tissues. (a) Protein extracts were prepared from normal thyroid, gallbladder (GB), appendix, kidney, spleen, skin, and small intestine, followed by western blot analysis using Hep Par 1 antibody. The Hep Par 1 antibody detects a 165 kDa band in the small intestine tissue only. Actin was used to serve as an internal control. (b) Immunohistochemical staining of normal small intestine tissue with Hep Par 1 antibody (1:50 dilution; original magnification × 200).
Mass Spectrometry Identifies p165 as CPS1
Since the p165 protein appeared to be abundant on the basis of immunoreactivity in western blots, we performed immunoprecipitation using Hep Par 1 antibody for the purpose of protein purification. After immunoprecipitation, the protein was eluted and resolved in SDS-PAGE. The gel was then stained with Coomassie blue. As shown in Figure 4, a distinctive p165 kDa band was readily visible. The p165 band was excised from the gel and used for protein identification.
Immunoprecipitation of liver extracts and frozen fresh liver tissue. Liver extract was incubated with Hep Par 1 antibody, followed by protein A selection. The protein elute was resolved in 10% SDS-PAGE and stained with Coomassie blue. The 165 kDa protein band was then excised from the gel and used for mass spectrometry analysis. All the four lanes were loaded with same liver tissue extract after immunoprecipitation. The amount difference reflects variations during multiple steps of immunoprecipitation.
LC–MS/MS analysis was then performed, followed by database search against NCBI_nr using MASCOT for the tryptic digests of the protein from the distinct band within the SDS-PAGE gel. The sequence and mass data of a total 116 peptide fragments matched a protein coded as IPI00011062, which is CPS1, with a high matching score of 2016 (see Table 1). The protein matching score is the sum of the unique ion scores, which is based on the absolute probability (P) that the observed match between the experimental data and the database sequence is a random event. The reported score is −10Log (P). The nominal molecular mass for the identified protein (Mr) is 165 ,975 kDa and the calculated pI is 6.30; the molecular mass is close to the experimental range of the gel band displayed in the SDS-PAGE (Figure 4). The identified peptide sequences covered 36% of the total amino-acid sequence of carbamoyl-phosphate synthase 1. These findings identified the Hep Par 1 antigen (p165) to be carbamoyl-phosphate synthase 1 (CPS1) with a very high degree of probability.
To further confirm that CPS1 is the antigen detected by Hep Par 1 antibody, we performed immunoprecipitation analysis. We precipitated liver protein extract with either Hep Par 1 antibody (Figure 5a) or anti-CPS1 antibody (Figure 5b). The eluted immunoprecipitate products were resolved in SDS-PAGE, followed by transferring to membrane and incubating the membrane with anti-CPS1 antibody (Figure 5a) or Hep Par 1 antibody (Figure 5b). As shown in Figure 5, anti-CPS1 specifically reacted with the Hep Par 1-precipitated 165 kDa protein (Figure 5a) and Hep Par 1 reacted with the 165 kDa protein pulled down by anti-CPS1. Moreover, we stained a hepatocellular carcinoma tissue section with anti-CPS1 antibody, knowing that this specific liver tumor was negative for Hep Par 1 immunoreactivity. As shown in Figure 5c, the tumor cells are negative for CPS1 and the residual normal hepatocytes are strongly positive for CPS1. The granular cytoplasmic staining pattern in the residual hepatocytes is similar to that of Hep Par 1 antibody staining pattern.
Hep Par 1 antibody and CPS1 recognize p165 protein. (a) Hep Par 1 immunoprecipitated protein reacts with anti-CPS1 antibody. Liver extract was incubated with control immunoglobulin (lanes 1 and 2) or Hep Par 1 antibody (lanes 3 and 4) and protein A selection. The protein elute was then analyzed by western blot using an anti-CPS1 polyclonal antibody. (b) Liver extract was first incubated with control immunoglobulin (lane 1) and anti-CPS1 antibody (Lane 2), followed by protein A selection and western blot analysis with Hep Par 1 antibody (lane 2). (c) Immunohistochemical staining for CPS1 of hepatocellular carcinoma tissue with negative Hep Par 1 staining. The tumor cells are negative for anti-CPS1; residual normal hepatocytes in the surrounding cirrhotic liver are strongly positive for Hep Par 1 and anti-CPS1. Magnification, × 400.
As noted in the Introduction, Hep Par 1 antibody also reacts with some non-liver cancer cells, especially the ‘hepatoid component’. We therefore examined the CPS1 staining pattern in two types of tumors available to us: a gastric adenocarcinoma with hepatoid differentiation, and several yolk sac tumors of the ovary. As shown in Figure 6a, the hepatoid component of the gastric carcinoma was strongly positive for both Hep Par 1 and CPS1. Similarly, the focal Hep Par 1-positive tumor cells in the one yolk sac tumor exhibiting ‘hepatoid’ features were also positive for CPS1. The two other yolk sac tumors exhibited negative immunoreactivity for both Hep Par 1 and CPS1 (data not shown).
Hep Par 1 and CPS1 have similar immunostaining pattern in non-liver tumors. (a) A ‘hepatoid’ gastric carcinoma shows immunoreactivity with both Hep Par 1 and anti-CPS1 antibodies (magnification, × 200). (b) A yolk sac tumor showing same group of cells with both CPS1 and Hep Par 1 immunoreactivity (magnification, × 400).
Collectively, these experiments firmly establish the connection between Hep Par 1 and CPS1.
CPS1 Expression is Suppressed at the Transcriptional Level in Human Hepatocellular Carcinoma Cell Lines
Figure 2 shows that Huh-7 cells are negative for Hep Par 1. We then examined the CPS1 expression in other hepatocellular carcinoma cell lines, HepG2 and LH86. As shown in Figure 7a, none of the three hepatocellular carcinoma cell lines express CPS1, while cultured human primary hepatocytes strongly express the CPS1 protein. Total RNA was then extracted from both primary hepatocytes and Huh-7 cells, followed by RT-PCR analysis. As shown in Figure 7b, CPS1 RNA is readily detected in hepatocytes, but not Huh-7 cells. To rule out the possibility of genomic deletion of CPS1 gene in Huh-7 cells, genomic DNA was obtained from both hepatocytes and Huh-7 cells. As shown in Figure 7c, the CPS1 gene is present in both cells. These data indicate that suppression of CPS1 expression occurs at the RNA transcriptional level. The fact that both CPS1 mRNA and protein are expressed in primary cultured human hepatocytes excludes culture conditions per se as the reason for downregulation of CPS1 expression.
Cultured hepatocellular carcinoma cell lines do not have CPS1 expression. (a) Western blot analysis of cellular extracts from cultured cells using Hep Par 1 antibody. Lane 1: primary hepatocytes in culture; lanes 2–4: three different clones of Huh-7 cells; lane 5, HepG2 cells; lane 6: LH86 cells. The figure shows that CPS1 is expressed only in primary cultured hepatocytes. Actin serves as an internal control. (b) CPS1 mRNA expression in normal hepatocytes and Huh-7 cells using RT-PCR analysis; only primary cultured hepatocytes express CPS1 mRNA. (c) CPS1 gene in primary cultured hepatocytes and Huh-7 cells using PCR analysis of genomic DNA. Gene status is intact in both primary cultured hepatocytes and Huh-7 cells.
DISCUSSION
Hep Par 1 antibody is widely used in surgical pathology practice; however, the protein it reacts with is unknown despite a decade-and-a-half of clinical use. Our goal was to identify this protein. After confirming the presence of antigen that reacts with Hep Par 1 antibody in fresh liver tissue, we performed immunoprecipitation, which allowed us to identify a 165 kDa protein in the liver tissue. After purification of the p165 protein, we analyzed the protein using a standard mass spectrometry method. As shown in Table 1, the majority of digested peptides correspond to CPS1.
To more definitively establish this link, we performed immunoprecipitation–western blot analysis, whereby protein immunoprecipitated with Hep Par 1 was immunoreactive on western blot with anti-CPS1, and protein immunoprecipitated with anti-CPS1 was immunoreactive on western blot with Hep Par 1 (Figure 5). Immunohistochemical staining of non-liver tumors immunoreactive for Hep Par 1 (one gastric carcinoma and one yolk sac tumor) also demonstrated immunoreactivity for CPS1 in the same region of the tumors (Figure 6). We cannot totally exclude the possibility that Hep Par 1 antibody may also bind to other proteins. However, the evidence in this report supports the notion that Hep Par 1 predominantly binds to CPS1.
CPS1 is a rate-limiting enzyme in urea cycle.7, 21, 22 Converting ammonia to urea is one of the essential functions of the liver. CPS1 is an abundant hepatocellular protein and predominantly localizes in the mitochondrion, consistent with the granular immunohistochemical staining pattern of the Hep Par 1 antibody. Transcription of the CPS1 gene is subject to physiological regulation23, 24, 25 and a hepatotoxic dose of acetaminophen can inhibit CPS1 activity.26 CPS1 deficiency is a genetic syndrome that severely affects development due to increased level of ammonia.27 In some of these patients, the liver may show focal glycogenosis.28
It has been reported that CPS1 can be released from the hepatocytes during liver injury.29, 30 However, the clinical application of the elevated serum CPS1 remains to be determined. More importantly, the connections between altered CPS1 expression in human hepatocytes in disease conditions, especially hepatocarcinogenesis, has not been explored. It is possible that CPS1 alteration may be a surrogate marker to reflect the function of normal hepatocytes or cancerous ‘well-differentiated’ hepatocytes. The identification of CPS1 as the antigen for Hep Par 1 antibody now justifies investigation of the potential role of CPS1 in the pathobiology of the liver, especially hepatocarcinogenesis. In particular, it will be valuable to determine whether CPS1 expression plays a causal role in the differentiation state of tumorous hepatocytes and non-liver cells, or is instead only a surrogate marker for hepatocellular differentiation. Even in the latter instance, determining what regulates CPS1 gene expression may be a valuable future line of investigation.
There are two additional interesting observations in our study: (1) the presence of CPS1 in small intestinal epithelium; (2) the absence of CPS1 in a cultured hepatocellular carcinoma cell lines with the suppression occurring at the transcriptional level. Since CPS1 is an enzyme in the urea cycle, the intriguing question is whether small intestinal epithelial cells contain a functional urea cycle, or whether the presence of CPS1 denotes a previously unappreciated nitrogen-processing pathway in small intestinal epithelium, as suggested by studies using pig enterocytes.31, 32 Future experiments should be considered to examine the other urea cycle enzymes in the small intestinal epithelium. Importantly, examination of ammonia metabolism within the small intestine epithelium could provide important insights into intestinal epithelial physiology, particularly given a recent report that CPS1 polymorphisms constitute a risk condition for neonatal necrotizing enterocolitis.33
The absence of CPS1 mRNA and protein in the three hepatocellular carcinoma cell lines suggests that CPS1 is only expressed in more differentiated HCC, recognizing that expression of highly differentiated cellular proteins may be downregulated in immortalized cell lines. In fact, clinical studies have demonstrated that Hep Par 1 does not appear to react with poorly differentiated HCC.34, 35 A recent study has shown that CPS1 gene transcription is downregulated in liver cancer tissues.36 However, the absence of CPS1 in the cancer cell lines cannot be explained entirely by the hepatocellular differentiation state. It is known that all three cell lines express abundant levels of albumin and α-1 antitrypsin, indicating that many liver-specific transcription factors are present in these cell lines. The total absence of CPS1 in these cell lines is highly suggestive of other regulatory mechanisms of CPS1 gene expression control, such as epigenetic regulation. Future experimentation is needed to reveal the mechanisms.
The pathophysiological or functional significance of CPS1 in cancer cell differentiation and development remains to be determined. Given that disposal of excess nitrogen via the urea cycle is essential for life, underexpression of this key enzyme of the urea cycle raises intriguing questions about the nitrogen balance of malignant tumors. For the most part, only HCC express Hep Par 1 (and, hence, CPS1). We do not know whether function of the urea cycle has any role to play in carcinogenesis, or whether such function may prove to be of benefit for treatment of these recalcitrant tumors.
In conclusion, our study has identified CPS1 as the antigen reacting with the Hep Par 1 antibody. CPS1 is expressed in human hepatocytes and small intestinal epithelium, with silenced expression in human hepatocellular carcinoma cell lines at the gene transcriptional level. Our findings should promote further investigations of CPS1 in the context of underlying liver diseases and hepatocarcinogenesis.
References
Minervini MI, Demetris AJ, Lee RG, et al. Utilization of hepatocyte-specific antibody in the immunocytochemical evaluation of liver tumors. Mod Pathol 1997;10:686–692.
Wennerberg AE, Nalesnik MA, Coleman WB . Hepatocyte paraffin 1: a monoclonal antibody that reacts with hepatocytes and can be used for differential diagnosis of hepatic tumors. Am J Pathol 1993;143:1050–1054.
Lamps LW, Folpe AL . The diagnostic value of hepatocyte paraffin antibody 1 in differentiating hepatocellular neoplasms from nonhepatic tumors: a review. Adv Anat Pathol 2003;10:39–43.
Zimmerman RL, Burke MA, Young NA, et al. Diagnostic value of hepatocyte paraffin 1 antibody to discriminate hepatocellular carcinoma from metastatic carcinoma in fine-needle aspiration biopsies of the liver. Cancer 2001;93:288–291.
Chu PG, Ishizawa S, Wu E, et al. Hepatocyte antigen as a marker of hepatocellular carcinoma: an immunohistochemical comparison to carcinoembryonic antigen, CD10, and alpha-fetoprotein. Am J Surg Pathol 2002;26:978–988.
Fasano M, Theise ND, Nalesnik M, et al. Immunohistochemical evaluation of hepatoblastomas with use of the hepatocyte-specific marker, hepatocyte paraffin 1, and the polyclonal anti-carcinoembryonic antigen. Mod Pathol 1998;11:934–938.
Hall LM, Johnson RC, Cohen PP . The presence of carbamyl phosphate synthetase in intestinal mucosa. Biochim Biophys Acta 1960;37:144–145.
Maitra A, Murakata LA, Albores-Saavedra J . Immunoreactivity for hepatocyte paraffin 1 antibody in hepatoid adenocarcinomas of the gastrointestinal tract. Am J Clin Pathol 2001;115:689–694.
Lugli A, Tornillo L, Mirlacher M, et al. Hepatocyte paraffin 1 expression in human normal and neoplastic tissues: tissue microarray analysis on 3,940 tissue samples. Am J Clin Pathol 2004;122:721–727.
Kakar S, Muir T, Murphy LM, et al. Immunoreactivity of Hep Par 1 in hepatic and extrahepatic tumors and its correlation with albumin in situ hybridization in hepatocellular carcinoma. Am J Clin Pathol 2003;119:361–366.
Trompetas V, Varsamidakis N, Frangia K, et al. Gastric hepatoid adenocarcinoma and familial investigation: does it always produce alpha-fetoprotein? Eur J Gastroenterol Hepatol 2003;15:1241–1244.
Fan Z, van de Rijn M, Montgomery K, et al. Hep par 1 antibody stain for the differential diagnosis of hepatocellular carcinoma: 676 tumors tested using tissue microarrays and conventional tissue sections. Mod Pathol 2003;16:137–144.
Villari D, Caruso R, Grosso M, et al. Hep Par 1 in gastric and bowel carcinomas: an immunohistochemical study. Pathology 2002;34:423–426.
Zhang J, Parwani AV, Ali SZ . Hepatocyte paraffin 1 immunoexpression in esophageal brush samples. Cancer 2005;105:304–309.
Zhu H, Dong H, Eksioglu E, et al. Hepatitis C virus trigger apoptosis of a newly developed hepatoma cell line through antiviral defense system. Gastroenterolgy 2007;133:1649–1659.
Shang XZ, Zhu H, Lin K, et al. Stabilized beta-catenin promotes hepatocyte proliferation and inhibits TNFalpha-induced apoptosis. Lab Invest 2004;84:332–341.
Zhu H, Zhao H, Collins CD, et al. Gene expression associated with interferon alfa antiviral activity in an HCV replicon cell line. Hepatology 2003;37:1180–1188.
Zhu H, Nelson DR, Crawford JM, et al. Defective Jak-Stat activation in hepatoma cells is associated with hepatitis C viral IFN-alpha resistance. J Interferon Cytokine Res 2005;25:528–539.
LeCluyse EL, Alexandre E, Hamilton GA, et al. Isolation and culture of primary human hepatocytes. Methods Mol Biol 2005;290:207–229.
Chu PG, Jiang Z, Weiss LM . Hepatocyte antigen as a marker of intestinal metaplasia. Am J Surg Pathol 2003;27:952–959.
Metzenberg RL, Marshall M, Cohen PP . Purification of carbamyl phosphate synthetase from frog liver. J Biol Chem 1958;233:102–105.
Jones ME . Amino acid metabolism. Annu Rev Biochem 1965;34:381–418.
Nyunoya H, Broglie KE, Widgren EE, Lusty CJ . Characterization and derivation of the gene coding for mitochondrial carbamyl phosphate synthetase I of rat. J Biol Chem 1985;260:9346–9356.
Adcock MW, O'Brien WE . Molecular cloning of cDNA for rat and human carbamyl phosphate synthetase I. J Biol Chem 1984;259:13471–13476.
Tillman JB, Dhahbi JM, Mote PL, et al. Dietary calorie restriction in mice induces carbamyl phosphate synthetase I gene transcription tissue specifically. J Biol Chem 1996;271:3500–3506.
Gupta S, Rogers LK, Taylor SK, et al. Inhibition of carbamyl phosphate synthetase-I and glutamine synthetase by hepatotoxic doses of acetaminophen in mice. Toxicol Appl Pharmacol 1997;146:317–327.
Gelehrter TD, Snodgrass PJ . Lethal neonatal deficiency of carbamyl phosphate synthetase. N Engl J Med 1974;290:430–433.
Badizadegan K, Perez-Atayde AR . Focal glycogenosis of the liver in disorders of ureagenesis: its occurrence and diagnostic significance. Hepatology 1997;26:365–373.
Struck J, Uhlein M, Morgenthaler NG, et al. Release of the mitochondrial enzyme carbamoyl phosphate synthase under septic conditions. Shock 2005;23:533–538.
Crouser ED, Julian MW, Huff JE, et al. Carbamoyl phosphate synthase-1: a marker of mitochondrial damage and depletion in the liver during sepsis. Crit Care Med 2006;34:2439–2446.
Wu G . Urea synthesis in enterocytes of developing pigs. Biochem J 1995;312 (Part 3):717–723.
Davis PK, Wu G . Compartmentation and kinetics of urea cycle enzymes in porcine enterocytes. Comp Biochem Physiol B Biochem Mol Biol 1998;119:527–537.
Moonen RM, Paulussen AD, Souren NY, et al. Carbamoyl phosphate synthetase polymorphisms as a risk factor for necrotizing enterocolitis. Pediatr Res 2007;62:188–190.
Kumagai I, Masuda T, Sato S, et al. Immunoreactivity to monoclonal antibody, Hep Par 1, in human hepatocellular carcinomas according to histopathological grade and histological pattern. Hepatol Res 2001;20:312–319.
Sugiki T, Yamamoto M, Aruga A, et al. Immunohistological evaluation of single small hepatocellular carcinoma with negative staining of monoclonal antibody hepatocyte paraffin 1. J Surg Oncol 2004;88:104–107.
Kinoshita M, Miyata M . Underexpression of mRNA in human hepatocellular carcinoma focusing on eight loci. Hepatology 2002;36:433–438.
Acknowledgements
This work is partially supported by a clinical research grant from the Department of Pathology, University of Florida and a grant from NIDDK (DK02958). We thank members of Liu laboratory and Robin Foss for technical support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Butler, S., Dong, H., Cardona, D. et al. The antigen for Hep Par 1 antibody is the urea cycle enzyme carbamoyl phosphate synthetase 1. Lab Invest 88, 78–88 (2008). https://doi.org/10.1038/labinvest.3700699
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.3700699
Keywords
This article is cited by
-
Caspase-7 activates ASM to repair gasdermin and perforin pores
Nature (2022)
-
Hepatocyte paraffin 1 and arginase-1 are effective panel of markers in HBV-related HCC diagnosis in fine-needle aspiration specimens
BMC Research Notes (2020)
-
Spatially resolved analysis of FFPE tissue proteomes by quantitative mass spectrometry
Nature Protocols (2020)
-
Kinetic modelling of quantitative proteome data predicts metabolic reprogramming of liver cancer
British Journal of Cancer (2020)
-
Extracellular vesicles in liver disease and potential as biomarkers and therapeutic targets
Nature Reviews Gastroenterology & Hepatology (2017)