Abstract
Synovial sarcomas are soft-tissue tumors predominantly affecting children and young adults. They are molecular-genetically characterized by the SYT-SSX fusion gene generated from chromosomal translocation t(X; 18) (p11.2; q11.2). When we screened new gene products that interact with SYT or SSX proteins by yeast two-hybrid assay, we found that mSin3A, a component of the histone deacetylase complex, interacts with SYT but not with SSX. These results were confirmed by mammalian two-hybrid and pull-down assays. Analyses with sequential truncated proteins revealed a main mSin3A-interaction region on the SYT amino-terminal 93 amino acids, and another one on the region between 187th amino acid and break point. In luciferase assay, mSin3A repressed the transcriptional activity of reporter promoter mediated by SYT and hBRM/BRG1. Our results suggest that the histone deacetylase complex containing mSin3A may regulate the transcriptional activation mediated by SYT.
Similar content being viewed by others
Main
Synovial sarcomas typically occur in the para-articular regions of adolescents and young adults, and account for about 10% of all soft-tissue sarcomas. They are considered high-grade sarcomas because they lead to death in at least 25% of patients within 5 years of diagnosis despite advances in treatment.1 Synovial sarcomas are characterized cytogenetically by the presence of a chromosomal translocation t(X;18)(p11.2;q11.2).2 Using positional cloning strategies, it was found that the SYT gene on chromosome 18 was translocated to one of two closely related SSX genes, SSX1 and SSX2, on chromosome X.3
The SYT gene is ubiquitously expressed in the early stages of embryonic development, and detected in several organs, most markedly the heart, kidney and testis, in the adult mouse.4 Using immunofluorescence analysis, it has been shown that the SYT protein exhibits a punctated pattern in nucleus.5 Since no colocalization of SYT with any nuclear bodies could be found by these researchers, it was suggested that SYT might be associated with a new class of subnuclear domains.5, 6 Four putative src-homology binding domains and two annexin-like direct repeats were exhibited in the SYT protein.3, 4 Recently, two domains were also defined in SYT proteins: one is the SYT amino (N)-terminal homology (SNH) domain, which shows homology to the predicted proteins of EST clones derived from a wide variety of species, and the other is the carboxy (C)-terminal QPGY domain, which is rich in glutamine, proline, glycine and tyrosine.7, 8 The QPGY domain was shown to activate the transcription of a reporter gene when fused to a DNA binding domain.6, 8 Since the SYT protein lacks obvious DNA binding domains, it is thought to be a transcriptional coactivator.6 It has also been reported that SYT is associated with a chromatin remodeling factor hBRM/hSNF2 alpha protein,8 a transcriptional coactivator p300 protein,9 and a putative transcriptional factor AF10.7 Recent studies have also shown that the SYT gene produces a splicing variant that contains 31 amino acids in the QPGY domain of conventional SYT,10, 11 and that the splicing isoform of the SYT-SSX fusion protein accelerate the transcriptional activity and cell proliferation.12
SSX proteins contain an acidic C-terminal tail and a Kruppel-associated box (KRAB) in their N-terminus.13, 14 The KRAB-domain was first reported as a transcriptional repressor domain in the Kruppel gene product of Drosophila. Functional studies have shown that the acidic C-terminus of SSX carries another repressor domain,6, 8 which is referred as SSX-RD.14 SSX proteins are nuclear proteins that exhibit a diffuse staining pattern in the nucleus.5, 6 In addition, the SSX proteins showed colocalization with several members of the Polycomb group proteins which are associated with transcriptional repression.14
In most SYT-SSX fusion proteins, C-terminal eight amino acids of SYT are replaced by C-terminal 78 amino acids of SSX.3, 13, 15 Recently, six genes have been identified as members of the SSX gene family,16, 17, 18 and the translocation of the SYT and SSX4 genes was detected in a case of synovial sarcoma.19
Acetylation of histone H3 and H4 is associated with the activation of gene expression.20, 21 Some transcriptional coactivators such as p300/CBP, have histone acetyltransferase activity. On the other hand, histone deacetylase (HDAC) functions in the transcriptional corepressor complex.22 HDAC1 and HDAC2 are associated with mSin3A, N-CoR, SMRT and some other components to form mSin3A/HDAC complexes.23 The complexes act on chromatin through transcriptional factors, corepressors and methyl-CpG binding proteins, leading to repression of gene transcription.24
In the present study, we performed a yeast two-hybrid assay to identify new proteins, which interact with SYT or SSX protein, and found that mSin3A binds to the SYT protein, and that mSin3A represses the transcriptional activity mediated by SYT.
Materials and methods
Complementary DNA Synthesis
The coding regions of the human SYT, SSX1, SSX2, SYT-SSX1 and SYT-SSX2 cDNAs were amplified by polymerase chain reaction (PCR) with human cDNAs (derived from synovial sarcoma or testis) and a suitable pair of the following primers: SYT S1-sense 5′-CTG AAT TCA TGG GCG GCA ACA TGT C and antisense 5′-TTC ACT GCT GGT AAT TTC CAT ACT, SSX sense 5′-GGT GCC ATG AAC GGA GAC GA and antisense 5′-GGA GTT ACT CGT CAT CTT CCT C. The same primers were used for amplification of the SSX1 and SSX2 cDNAs, and the two products were confirmed by sequencing. mSin3A cDNA encompassing codon 1 and 758 of the mSin3A gene was amplified by PCR with primers 5′-AGG ATC CAT GAA GCG ACG GTT GGA TGA CC and 5′-TCT CGA GAT AGA TGC TCT CGA TCT CAT T.25 Human complementary DNA (cDNA) encompassing codon 194 and 657 of the SMRT gene was amplified by PCR with primers 5′-AGA ATT CAA AGG CAT TCC CAG CAC AC and 5′-ACT CGA GCT TGC TGG ACG AAA GGC TG.26 Human N-CoR cDNA encompassing codon 1019 and 2061 of the N-CoR gene was made by PCR with primers 5′-TGA ATT CCC TGA AGG CGT TCG GCT TC and 5′-ACT CGA GGG AGT CTG CGA GGA AAC TTG.27 Human HDAC1 full-length cDNA was amplified by PCR with primers 5′-TGG ATC CAA GAT GGC GCA GAC GCA GG and 5′-AGT CGA CTC AGG CCA ACT TGA CCT CCT C. The amplified cDNAs were cloned into pBluescript plasmid, and sequenced with a BigDye sequencing kit and an ABI3100 sequencer (Applied Biosystems, Foster City, CA, USA). A series of truncated SYT cDNA was constructed by PCR with following primers: SYT S1-sense and BP-antisense 5′-AAC TCG AGC TAC TGG TCA TAT CCA TAA GG for SYT (1-BP) cDNA, SYT S1 and antisense 5′-TTC TCG AGT TAA CTC ATT GTC ATC TGA TTC TG for SYT (1–186) cDNA, SYT S1-sense and antisense 5′-TTC TCG AGT TAC CCT CCA GGA CCC ATA GG for SYT (1–93) cDNA, SYT sense 5′-CCG AAT TCA TGG GAG GGA TGA ATC AGA GCG GC and BP-antisense for SYT (94-BP) cDNA, and SYT sense 5′-CCG AAT TCA TGG GGG GTC AGG GAC AAC CAA TGG GAA AC and BP-antisense for SYT (187-BP) cDNA.
Yeast Two-Hybrid Assay
The human SYT, SSX1, SSX2, SYT-SSX1 and SYT-SSX2 cDNAs were inserted into the pBTM116 plasmid to produce fusion proteins with the LexA DNA binding domain in frame. SYT-SSX2T cDNA, a truncated type of SYT-SSX2 gene, was made by digesting the SYT-SSX2 cDNA with Sma I on the SSX2 and Pst I on the 3′ cloning site of pBTM116 plasmid, to make fusion protein containing the N-terminal region of SYT and the 111–143 amino acid region of SSX2. Human mSin3A, N-CoR, SMRT and HDAC1 cDNAs were inserted into the pACT2 plasmid to produce fusion proteins with the GAL4 activation domain in frame. L40 yeast cells (MATa ade2 gal4 gal80 his3-Δ200 leu2-3,113 trpl-Δ901 ura3-52 URA3::(lexAop)8-lacZ LYS2::(lexAop)4-HIS3) were transformed with pBTM116 plasmids bearing the TRP1 gene and pACT2 plasmids bearing the LEU2 gene, and selected on synthetic medium plates without His, Trp and Leu at 30°C for 2 days. The candidate colonies on the selective plates were examined by filter assay for β-galactosidase according to the Clontech protocol (Clontech Laboratory, Palo Alto, CA, USA). In brief, colonies were transferred onto Whatman filters, frozen in liquid nitrogen and thawed to permeabilize the yeast cells. The colonies on the filters were incubated by immersion in Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, pH 7.0) including 39 mM β-mercaptoethanol and 0.4 mg/ml 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside(X-gal) at 30°C. For liquid culture assays of β-galactosidase, multiple colonies were scraped off from each plate and transferred to 1 ml of distilled water. After thoroughly mixing, the absorbance of each suspension was read at 600 nm (OD600). The cell suspension (0.1 ml), Z buffer (0.7 ml), 0.1% SDS (50 μl) and chloroform (50 μl) were mixed and vortexed for 30 s. Assays for β-galactosidase were initiated by the addition of 160 μl of 4 mg/ml o-nitrophenyl β-D-galactopyranoside at 30°C. Reactions were terminated by the addition of 0.4 ml of 1 M Na2CO3. Reaction mixtures were centrifuged for 1 min, and the absorbance at 420 nm (OD420) was measured. β-Galactosidase activity in Miller units was calculated according to the following formula: units=1000 × OD420/t × v × OD600 (where v is the volume (ml) of culture added to Z buffer, and t is the time in min).
Mammalian Two-Hybrid Assay
The human SYT, SSX1, SSX2, SYT-SSX1 and SYT-SSX2 cDNAs were inserted into the pCMV-BD plasmid to produce fusion proteins with the GAL4 DNA binding domain in frame. Human mSin3A cDNAs were inserted into the pCMV-AD plasmid to produce fusion proteins with the NF-kB activation domain in frame. Human embryonic kidney 293 cells were transfected with pCMV-BD plasmid (150 ng), pCMV-AD plasmid (150 ng), GAL4-luciferase reporter plasmid (pRF-luciferase, 100 ng) and Renilla luciferase internal control plasmid (pTK-hRG, 100 ng) by Effectene reagent (Qiagen, Hilden, Germany). Dual-luciferase activity was measured 48 h after transfection as described previously.28
Pull-Down Assay
35S-labeled proteins were generated using TnT T7 quick-coupled transcription/translation system (Promega, Madison, WI, USA) with mSin3A cDNA in pBluescript. GST-fused SYT proteins were purified with glutathione–sepharose 4B beads (Amersham Biosciences, Piscataway, NJ, USA) in lysis buffer (25 mM tris-HCl (pH 8.0), 150 mM NaCl, 0.5% NP40 and proteinase inhibitors) at 48 h after transfection of HEK293T cells with mammalian GST expression vectors containing SYT, SYT-SSX and truncated SYT (1-BP) cDNA. The purified proteins on the beads were incubated with 35S-labeled mSin3A protein at 4°C for 3 h. The beads were washed six times, and loaded onto 8% SDS-polyacrylamide gels. Radioactive signals on the dried gels were detected by Bioimaging Analyzer System (BAS2000, Fuji, Tokyo).
Luciferase Assays
N-terminal SYT cDNA coding 1–93 amino acids was inserted into the pCMV-BD plasmid to produce fusion proteins with the GAL4 DNA binding domain in frame (pCMV-BD-SYT-N). Expression plasmids, pCI-Neo-hBRM and pcDNA3.1(+)-BRG1, were kindly provided by Dr T Ohta (National Cancer Institute, Japan), and pCMX-mSin3A was kindly provided by Dr RM Evans (The Salk Institute for Biological Studies, San Diego, CA, USA). HEK293T cells in a 48-well titer plate were transfected with pCMV-BD-SYT-N (40 ng), pRF-luciferase (75 ng), pCI-Neo-hBRM/pcDNA3.1(+)-BRG1 (30 ng), pCMX-mSin3A (30 ng) and internal control plasmid pTK-hRG (25 ng) for dual-luciferase system. DNA amount for transfection was compensated with empty vectors.
Results
Screening of the Binding Proteins to SYT or SSX Proteins
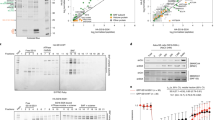
When we examined the interaction of SSX proteins with HDAC1, mSin3A, SMRT and N-CoR proteins using the yeast two-hybrid assay, we could not get any positive colonies, which showed protein–protein interaction (data not shown). On the other hand, when SYT was examined as a control, surprisingly we found that SYT can interact with mSin3A by β-galactosidase filter assay with a short incubation time (Figure 1a). As it has been reported that the SYT gene produces a splicing variant which contains 31 amino acids in the QPGY domain of conventional SYT,10, 11 we examined the interaction of the SYT variant with component proteins of mSin3A/HDAC complexes. The SYT variant also showed positive interaction with mSin3A. HDAC1, N-CoR and SMRT showed weak interaction with SYT and the variant SYT with a longer incubation time (data not shown), while control colonies containing empty vectors did not show any interaction with any of these four proteins. Liquid culture assay for β-galactosidase also showed interaction of SYT with mSin3A (Figure 1b).
Interaction of SYT or SYT-SSX with histone deacetylase complex proteins. (a) SYT and the variant with 31 amino acids (SYT(−31) and SYT(+31), respectively) showed interaction with mSin3A in yeast two-hybrid filter assay. (b) Intensity of the interaction is shown as β-galactosidase activity by yeast two-hybrid liquid culture assay. (c) SYT-SSX proteins showed less interaction with mSin3A.
Identification of the SYT Region Specific for Interaction with mSin3A
We constructed a series of SYT-truncated mutants to identify the region of SYT that interacts with mSin3A, and measured β-galactosidase activity to assess the interaction of the truncated SYT proteins and mSin3A (Figure 2). Surprisingly, when C-terminal eight amino acids were deleted, the truncated SYT proteins showed more intensive interaction with mSin3A. We defined the main interaction region for mSin3A on the N-terminal 1–93 amino acids of SYT, and another one on the region between the 187th amino acid and break point. There might be negative regulating regions for the interaction on C-terminal eight amino acids and 94–186 amino acids of SYT.
Identification of SYT region required for interaction with mSin3A. Truncated SYT proteins used for yeast two-hybrid liquid culture assay are shown at left, and β-galactosidase activity is shown at right. A black stripe indicates a 31 amino-acid insertion in the SYT variant, SYT(+31), and the truncated SYT variant, SYT(+31)(1-BP). Arrows show break point. Error bars indicate the standard deviation obtained from three independent assays.
Interaction of SYT-SSX Fusion Proteins with mSin3A
To clarify the interaction of mSin3A and the SYT-SSX oncoprotein, which is encoded on the fusion gene generated by the translocated chromosome t(18; X) found in synovial sarcomas, we constructed plasmids carrying SYT-SSX1 and SYT-SSX2 cDNA, and examined their interaction with mSin3A (Figures 1c and 3). SYT-SSX1 and SYT-SSX2 showed remarkably less β-galactosidase activity in the presence of mSin3A in yeast two-hybrid assay. Furthermore, we performed a mammalian two-hybrid analysis with human embryonic kidney 293 cells to confirm SYT interaction with mSin3A in human cells. SYT and the variant SYT with 31 amino acids showed positive interaction with mSin3A, while SYT-SSX proteins showed the lower level of luciferase activity in the presence of mSin3A (Figure 4). To define the inhibitory region on SSX, we constructed the truncated SYT-SSX plasmid in which the SSX C-terminal region (144–188 amino acids) was lost, and examined the interaction with mSin3A in yeast two-hybrid system (Figure 3). The β-galactosidase activity was restored by using the truncated SYT-SSX. The low activity of β-galactosidase and luciferase in the presence of SYT-SSX and mSin3A was thought to be due to the repression activity of SSX-RD region (155–188 amino acids) on SSX C-terminus. Therefore, we examined the direct binding of mSin3A with SYT and SYT-SSX by pull-down assay (Figure 5). When GST-fused SYT-SSX protein was pulled down in the presence of in vitro-translated mSin3A, mSin3A showed the binding with SYT-SSX as well as SYT and the truncated SYT.
Interaction of SYT-SSX proteins with mSin3A. Intensity of the interaction is shown as β-galactosidase activity by yeast two-hybrid liquid culture assay. SYT-SSX1 and SYT-SSX2 proteins show less interaction with mSin3A. When the C-terminal region (144–188 amino acids) was removed, the β-galactosidase activity was restored using SYT-SSX2T. SYT(+31)-SSX 1 and 2 were composed of the N-terminal 1–410 amino-acid region of SYT(+31) and the C-terminal 111–188 amino-acid region of SSX1 and 2, respectively. SYT-SSX2T was composed of the N-terminal region of SYT and the C-terminal 111–143 amino acid region of SSX2. SSX-DR is located on 155–188 amino acids region of SSX. Error bars indicate the standard deviation obtained from three independent assays.
Mammalian two-hybrid assay for interaction of SYT, SSX and SYT-SSX proteins with mSin3A. Schematic diagrams of the expression constructs are shown at left, and luciferase activity is shown at right. Arrows show break point. Error bars indicate the standard deviation obtained from four independent assays.
Pull-down assay for SYT and mSin3A. GST-SYT, GST-SYT-SSX1 and GST-truncated SYT (1-BP) were purified from transiently transfected HEK293T cells with glutathione–sepharose beads and incubated with 35S-methionine-labeled in vitro translated mSin3A. Specific binding of mSin3A to GST-SYT, GST-SYT-SSX1 and GST-SYT(1-BP) were detected (lanes 4–6, respectively). Translated lysate without mRNA and GST empty vector were used as negative controls (lanes 1 and 3, respectively). Input, sample representing total input (0.6%) for each experiment.
Effect of mSin3A on Transcriptional Activity Mediated by SYT
To examine the function of mSin3A, SYT N-terminal region was fused to GAL4 DNA binding domain, and the effect of mSin3A on SYT was analyzed using luciferase assay with hBRM and BRG1 genes. The hBRM and BRG1 are chromatin remodeling factors that regulate the transcriptional activity, and SYT is known to interact with them.8, 29 The GAL4-SYT products showed an increased transcriptional activity of the reporter gene in the presence of hBRM/BRG1, and mSin3A repressed the transcriptional activity of reporter promoter mediated by SYT and hBRM/BRG1 (Figure 6).
Effect of mSin3A on the transcriptional activity mediated by SYT and hBRM/BRG1. The indicated expression vectors (hBRM, BRG1 and mSin3A) were transfected into HEK293T cells with GAL4-luciferase reporter plasmid (pRF-Luc) and GAL4-fused SYT N-terminal (1–93) plasmid, pCMV-BD-SYT-N. Transfection efficiency was normalized using dual-luciferase system. Error bars indicate the standard deviation obtained from three independent assays. GAL4-SYT-N, pCMV-BD-SYT-N.
Discussion
As SSX proteins are thought to be transcriptional corepressors, we hypothesized that SSX proteins might interact with mSin3A/HDAC complexes, which are associated with transcriptional repression. We performed a two-hybrid assay to determine whether this was indeed the case. Surprisingly, our results showed interaction of mSin3A with SYT, but not with SSX. We found that the interaction of SYT with mSin3A requires mainly the N-terminal 1–93 amino acid region of SYT, and secondarily the region between the 187th amino acid and break point. Interestingly, deletion of C-terminal eight amino acids from SYT resulted in more intensive interaction of SYT with mSin3A. Although the two-hybrid assay with SYT-SSX and mSin3A showed less transcriptional activity, positive interaction of them was shown by pull-down assay. The decrease of transcriptional activity in the two-hybrid assay is likely due to repression activity of SSX C-terminal region (SSX-RD) directly or indirectly through the SSX-binding proteins like the Polycomb group proteins.14 Recently, it was reported that SSX C-terminal region of SYT-SSX protein repressed the activity of reporter promoter which was produced by binding of SYT N-terminal region and BRM/hBRG1.29 This is a similar case to our results.
On the promoter/enhancer regions of the target genes, SYT proteins may bind transcriptional factors like AF10. As it has been reported that p300 protein binds with the region between 1 and 250 amino acids of SYT,9 and that BRM binds with a major binding site between 60 and 158 amino acids of SYT,8 SYT may bind competitively with these proteins for transcriptional activation and with mSin3A for transcriptional repression. The complex containing mSin3A may function as a negative regulator for the transcriptional activation mediated by SYT in the normal cells. As oligomerization of QPGY domain on SYT was showed recently,29 SYT-SSX may act dominantly in the presence of normal SYT through the heterodimer of SYT and SYT-SSX proteins in synovial sarcoma cells. The repression activity of SSX C-terminal region on SYT-SSX may affect dominantly on the transcriptional regulation of SYT by p300, hBRM, BRG1 and mSin3A.
It has been reported that some fusion proteins originating from chromosomal translocation are associated with histone acetylation or deacetylation. PML-RARα and AML-ETO fusion proteins produce abnormal mSin3A/HDAC complexes in hematopoietic malignant cells.30, 31 Retinoic acid receptor alpha, RARα, binds to the HDAC complex for transcriptional repression in the absence of the ligand, and binds to the HAT complex for transcriptional activation in the presence of the ligand.32 Recently, it has been shown that human T-cell leukemia virus type 1 (HTLV-1) Tax protein, which activates transcription from its long terminal repeat by interacting with p300/CBP and CREB, can negatively regulate the gene expression by binding with HDAC1.33 These cases have a similarity with the relationship of SYT and mSin3A shown in the present study. The mSin3A/HDAC complex might negatively regulate the transcriptional activation mediated by SYT which is associated with transcriptional coactivators such as p300. Our findings after long incubation showed that HDAC1, SMRT and N-CoR weakly interact with SYT by yeast two-hybrid assay (data not shown). These proteins may bind with SYT indirectly through endogenous SIN3, which is a yeast homologue of mSin3A.
SYT-SSX fusion gene has been detected in nearly 97% cases of synovial sarcomas.34 At present, there is no effective treatment for synovial sarcomas, and the 5-year overall survival rates for synovial sarcomas are 53% for the SYT-SSX1 type and 73% for the SYT-SSX2 type.35 Our results reveal the possibility that an association of SYT and deacetylation may be critical for tumorigenesis in synovial sarcomas. Proteins that are regulated by acetylation and deacetylation are known to include not only histone but also tumor suppressor proteins and transcriptional factors that participate in regulation of differentiation, proliferation and apoptosis.36 Further analyses of the protein–protein interaction of SYT-SSX will be required to shed light on the mechanism of tumorigenesis in synovial sarcomas, and to develop an effective cure for synovial sarcomas.
References
Lewis JJ, Antonescu CR, Leung DH, et al. Synovial sarcoma: a multivariate analysis of prognostic factors in 112 patients with primary localized tumors of the extremity. J Clin Oncol 2000;10:2087–2094.
Turc-Carel C, Dal Cin P, Limon J, et al. Translocation X;18 in synovial sarcoma. Cancer Genet Cytogenet 1986;23:93.
Clark J, Rocques PJ, Crew AJ, et al. Identification of novel genes, SYT and SSX, involved in the t(X;18) (p11.2;q11.2) translocation found in human synovial sarcoma. Nat Genet 1994;7:502–508.
de Bruijn DR, Baats E, Zechner U, et al. Isolation and characterization of the mouse homolog of SYT, a gene implicated in the development of human synovial sarcomas. Oncogene 1996;13:643–648.
dos Santos NR, de Bruijn DRH, Balemans M, et al. Nuclear localization of SYT, SSX and the synovial sarcoma-associated SYT-SSX fusion proteins. Hum Mol Genet 1997;6:1549–1558.
Brett D, Whitehouse S, Antonson P, et al. The SYT protein involved in the t(X;18) synovial sarcoma translocation is a transcriptional activator localized in nuclear bodies. Hum Mol Genet 1997;6:1559–1564.
de Bruijn DR, dos Santos NR, Thijssen J, et al. The synovial sarcoma associated protein SYT interacts with the acute leukemia associated protein AF10. Oncogene 2001;20:3281–3289.
Thaete C, Brett D, Monaghan P, et al. Functional domains of the SYT and SYT-SSX synovial sarcoma translocation proteins and co-localization with the SNF protein BRM in the nucleus. Hum Mol Genet 1999;8:585–591.
Eid JE, Kung AL, Scully R, et al. p300 interacts with the nuclear proto-oncoprotein SYT as part of the active control of cell adhesion. Cell 2000;102:839–848.
Brodin B, Haslam K, Yang K, et al. Cloning and characterization of spliced fusion transcript variants of synovial sarcoma: SYT/SSX4, SYT/SSX4v, and SYT/SSX2v. Possible regulatory role of the fusion gene product in wild type SYT expression. Gene 2001;268:173–182.
Tamborini E, Agus V, Mezzelani A, et al. Identification of a novel spliced variant of the SYT gene expressed in normal tissues and in synovial sarcoma. Br J Cancer 2001;84:1087–1094.
Morimoto Y, Ouchida M, Ozaki T, et al. Splicing isoform of SYT-SSX fusion protein accelerates transcriptional activity and cell proliferation. Cancer Lett 2003;199:35–43.
Crew AJ, Clark J, Fisher C, et al. Fusion of SYT to two genes, SSX1 and SSX2, encoding proteins with homology to the Kruppel-associated box in human synovial sarcoma. EMBO J 1995;14:2333–2340.
Lim F, Soulez M, Koczan D, et al. A KRAB-related domain and a novel transcription repression domain in proteins encoded by SSX genes that are disrupted in human sarcomas. Oncogene 1998;17:2013–2018.
de Leeuw B, Balemans M, Olde Weghuis D, et al. Identification of two alternative fusion genes, SYT-SSX1 and SYT-SSX2, in t(X;18)(p11.2;q11.2)-positive synovial sarcomas. Hum Mol Genet 1995;4:1097–1099.
de Leeuw B, Balemans M, Geurts van Kessel A . A novel Kruppel-associated box containing SSX gene (SSX3) on the human X chromosome is not implicated in t(X;18)-positive synovial sarcomas. Cytogenet Cell Genet 1996;73:179–183.
dos Santos NR, Torensma R, de Vries TJ, et al. Heterogeneous expression of the SSX cancer/testis antigens in human melanoma lesions and cell lines. Cancer Res 2000;60:1654–1662.
Gure AO, Tureci O, Sahin U, et al. SSX: a multigene family with several members transcribed in normal testis and human cancer. Int J Cancer 1997;72:965–971.
Skytting B, Nilsson G, Brodin B, et al. A novel fusion gene, SYT-SSX4, in synovial sarcoma. J Natl Cancer Inst 1999;91:974–975.
Braunstein M, Sobel RE, Allis CD, et al. Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Mol Cell Biol 1996;16:4349–4356.
Megee PC, Morgan BA, Mittman BA, et al. Genetic analysis of histone H4: essential role of lysines subject to reversible acetylation. Science 1990;247:841–845.
Ogryzko VV, Schiltz RL, Russanova V, et al. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 1996;87:953–959.
McKenna NJ, Lanz RB, O'Malley BW . Nuclear receptor coregulators: cellular and molecular biology. Endocr Rev 1999;20:321–344.
Nan X, Ng HH, Johnson CA, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 1998;393:311–312.
Dhordain P, Lin RJ, Quief S, et al. The LAZ3 (BCL-6) oncoprotein recruits a SMRT/mSIN3A/histone deacetylase containing complex to mediate transcriptional repression. Nucleic Acids Res 1998;26:4645–4651.
Dhordain P, Albagli O, Lin RJ, et al. Corepressor SMRT binds the BTB/POZ repressing domain of the LAZ3/BCL6 oncoprotein. Proc Natl Acad Sci USA 1997;94:10762–10767.
Huynh KD, Bardwell VJ . The BCL-6 POZ domain and other POZ domains interact with the co-repressors N-CoR and SMRT. Oncogene 1998;17:2473–2484.
Kukita A, Kukita T, Maeda H, et al. Osteoclast-derived zinc finger (OCZF) protein with POZ domain, a possible transcriptional regulator, is involved in osteoclastogenesis. Blood 1999;94:1987–1997.
Perani M, Ingram CJ, Cooper CS, et al. Conserved SNH domain of the proto-oncoprotein SYT interacts with components of the human chromatin remodelling complexes, while the QPGY repeat domain forms homo-oligomers. Oncogene 2003;22:8156–8167.
Lutterbach B, Westendorf JJ, Linggi B, et al. ETO, a target of t(8;21) in acute leukemia, interacts with the N-CoR and mSin3 corepressors. Mol Cell Biol 1998;18:7176–7184.
Wang J, Hoshino T, Redner RL, et al. ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC1 complex. Proc Natl Acad Sci USA 1998;95:10860–10865.
Lin RJ, Nagy L, Inoue S, et al. Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature 1998;391:811–814.
Ego T, Ariumi Y, Shimotono K . The interaction of HTLV-1 Tax with HDAC negatively regulates the viral gene expression. Oncogene 2002;21:7241–7246.
dos Santos NR, de Bruijn DR, Geurts van Kessel A . Molecular mechanisms underlying human synovial sarcoma development. Genes Chromosomes Cancer 2001;30:1–14.
Ladanyi M, Antonescu CR, Leung DH, et al. Impact of SYT-SSX fusion type on the clinical behavior of synovial sarcoma: a multi-institutional retrospective study of 243 patients. Cancer Res 2002;62:135–140.
Kelly WK, O'Connor OA, Marks PA . Histone deacetylase inhibitors: from target to clinical trials. Expert Opin Invest Drugs 2002;11:1695–1713.
Acknowledgements
We gratefully acknowledge Dr Tsutomu Ohta (National Cancer Institute, Tokyo, Japan) for his kind provision of hBRM/BRG1 expression plasmids, and Dr Ronald M Evans (The Salk Institute for Biological Studies, San Diego, CA, USA) for provision of mSin3A expression plasmid. This work was supported by Grant-in Aid from the Ministry of Education, Science, Sports and Culture of Japan to KS.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ito, T., Ouchida, M., Ito, S. et al. SYT, a partner of SYT-SSX oncoprotein in synovial sarcomas, interacts with mSin3A, a component of histone deacetylase complex. Lab Invest 84, 1484–1490 (2004). https://doi.org/10.1038/labinvest.3700174
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.3700174
Keywords
This article is cited by
-
Synovial Sarcoma: A Complex Disease with Multifaceted Signaling and Epigenetic Landscapes
Current Oncology Reports (2020)
-
Identification and analysis of CXCR4-positive synovial sarcoma-initiating cells
Oncogene (2016)
-
Synovial sarcoma is a gateway to the role of chromatin remodeling in cancer
Cancer and Metastasis Reviews (2015)
-
A phase II trial of panobinostat in patients with advanced pretreated soft tissue sarcoma. A study from the French Sarcoma Group
British Journal of Cancer (2013)
-
Genome-wide recruitment to Polycomb-modified chromatin and activity regulation of the synovial sarcoma oncogene SYT-SSX2
BMC Genomics (2012)